Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors
Figures
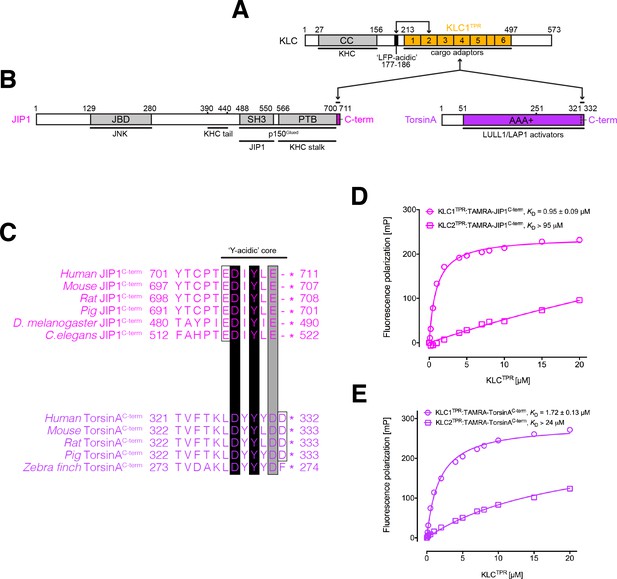
KLC1TPR binds Y-acidic peptides with low micromolar affinity.
(A) Schematic diagram of KLC. Residue numbers are for the human version of the KLC1 isoform. Important domains and protein regions are highlighted as well as key interacting partners. The LFP-acidic region (in black) engages in cis with KLCTPR (in gold, numbers correspond to the individual TPR repeats) contributing to the auto-inhibited state. (B) Schematic diagram of the JIP1 cargo adaptor and TorsinA proteins. They use their respective C-terminal regions to bind to KLC1TPR. The JIP1C-term (in magenta) is located immediately downstream of the PTB domain that is responsible for APP binding while TorsinAC-term (in violet) is an integral part of the AAA+ domain to which LULL1/LAP1 activators bind. (C) Multiple sequence alignment of JIP1C-term and TorsinAC-term peptides. Functionally unrelated JIP1 and TorsinA proteins share a conserved Y-acidic C-terminal region. Totally conserved Asp and Tyr residues (highlighted by black boxes), a conserved acidic residue (Glu/Asp highlighted by the grey box), and additional acidic residues (empty boxes) identify a common Y-acidic core. Residues upstream the Y-acidic core are not conserved. (D) Fluorescence polarization (FP) measurements indicate that TAMRA-labelled JIP1C-term binds with high affinity to KLC1TPR but not to the highly homologous KLC2TPR. (E) FP measurements show that TAMRA-labelled TorsinAC-term behaves similarly to JIP1C-term with a strong preference for KLC1TPR. The KD values reported in (D) and (E) refer to measurement carried out in buffer containing 150 mM NaCl. Error bars in the FP graphs are not visible as they are smaller than the size of the data point symbols.
-
Figure 1—supplement figure 1—source data 1
Fluorescence polarization data at varying NaCl concentrations as presented in panels A and B.
- https://doi.org/10.7554/eLife.38362.004
-
Figure 1—source data 1
Fluorescence polarization data for experiments presented in panels D and E.
- https://doi.org/10.7554/eLife.38362.006
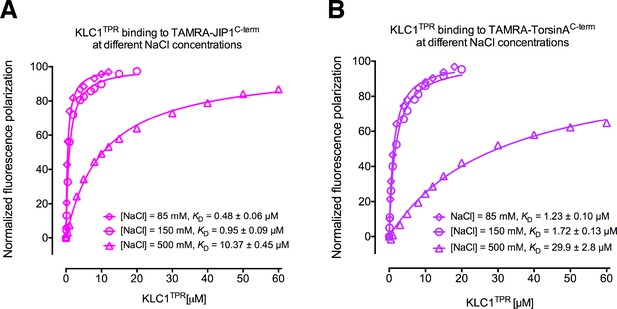
Binding affinity measurements between KLC1TPR and Y-acidic peptides at varying NaCl concentrations.
(A) FP measurements for the interaction with TAMRA-JIP1C-term. (B) FP measurements for the interaction with TAMRA-TorsinAC-term.
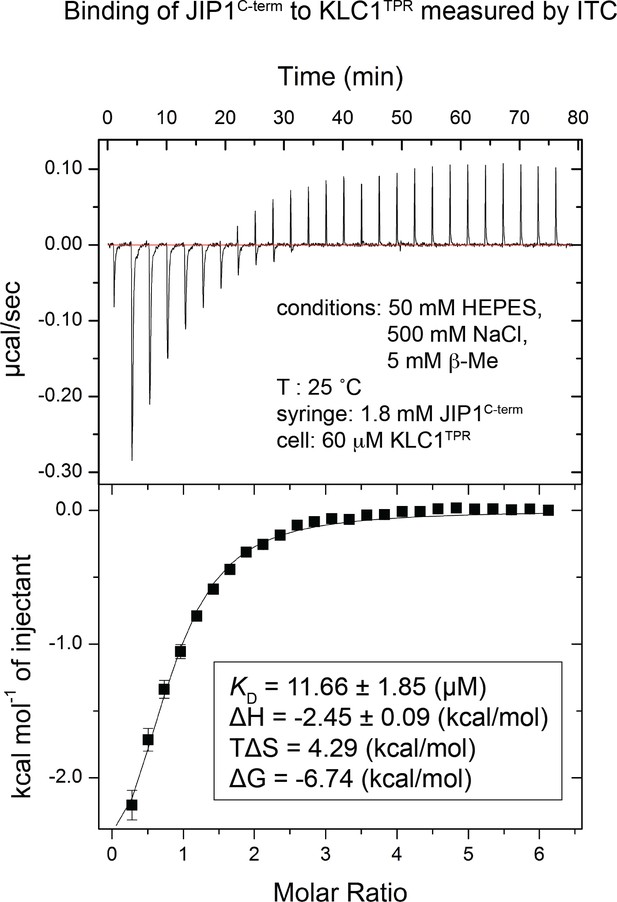
Binding of untagged JIP1C-term to KLC1TPR measured by isothermal titration calorimetry (ITC).
Untagged JIP1C-term (1.8 mM) in the syringe was titrated in the cell containing KLC1TPR (60 μM). The experiment was carried out at 25°C in 50 mM HEPES, pH 7.5, 500 mM NaCl, and 5 mM 2-mercaptoethanol. The binding constant and other thermodynamic parameters in the inset were calculated by fitting the integrated titration data assuming a single set of binding sites with N = 1 (1:1 stoichiometry) using the Origin software package (OriginLab).
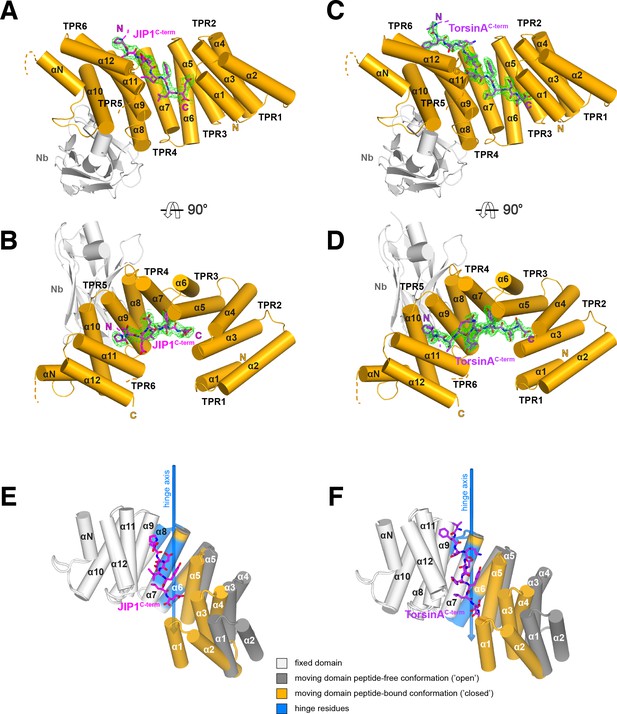
Overall structure of the complexes and conformational transition upon Y-acidic peptide recognition.
(A,B) KLC1TPR-JIP1C-term. Illustrated representations of the JIP1C-term peptide (displayed as a magenta stick model accompanied by its 2mFo-DFc electron density in green contoured at the 1.1σ level) bound to the KLC1TPR domain (gold) in two orthogonal views. The Nanobody (Nb) is shown in light grey. Helices α1 to α12 of the six TPR helix-turn-helix motifs and the non-TPR helix αN located between TPR5 and TPR6 are labeled. The flexible region between αN and α12 is indicated with a broken line while the unmodeled engineered linker connecting α12 to the JIP1C-term is not shown for clarity. Color-coded N and C labels indicate the N- and C-termini, respectively. Noncarbon elements are nitrogen and oxygen in dark blue and red, respectively. (C,D) KLC1TPR-TorsinAC-term. As (A,B) for the TorsinAC-term peptide colored in violet. (E,F) Conformational transition in the KLC1TPR domain upon recognition of Y-acidic peptides. Superposition of ligand-free KLC1TPR (PDB code 3NF1) and KLC1TPR-JIP1C-term (E) or KLC1TPR-TorsinAC-term (F) performed using the DynDom algorithm (Poornam et al., 2009). The analysis reveals the presence of an N-terminal 'moving' domain (highlighted in dark grey and gold for KLC1TPR and the KLC1TPR-Y-acidic complexes, respectively) and a C-terminal fixed domain (in light grey) with 'bending residues' shown in blue. The conformational transition requires a rotation of ∼30° degrees around the hinge axis represented by a blue arrow. Hinge residues are also highlighted in blue.
2mFo-DFc electron density of selected TPR helices in the KLC1TPR-TorsinAC-term complex.
(A) α1. (B) α3. Electron density is contoured at the 1.0σ level. Residues belonging to these helices are shown as stick representation with carbon, nitrogen, oxygen and sulfur atoms colored in gold, dark blue, red, and yellow, respectively. Helix α1 is less well defined compared to α3. The latter is representative of all other TPR helices.
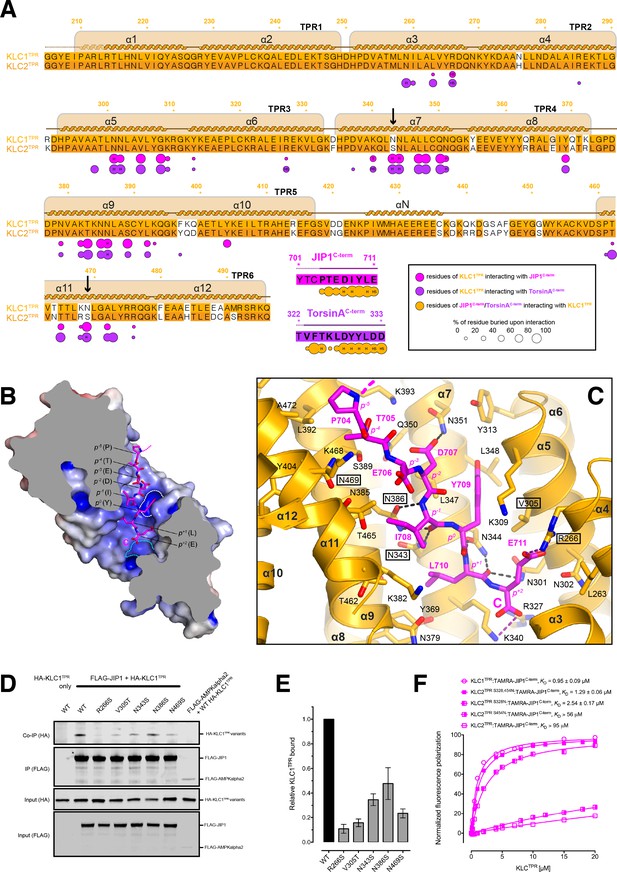
Molecular interface and cellular validation.
(A) Interface contact map for the KLC1TPR-JIP1C-term/TorsinAC-term complexes. The sequence of KLC2TPR is also shown for comparison. Colored circles indicate residues at the interface as indicated in the inset. Their radius is proportional to the buried area. The letter codes H and S highlight residues involved in hydrogen bonds and salt bridges, respectively. The black arrows highlight the only two residues making significant contacts with the Y-acidic peptides that are not conserved in KLC2TPR. Numbers are for KLC1TPR. (B) Sliced-surface view of the KLC1TPR domain with bound JIP1C-term. The molecular surface is colored according to its electrostatic potential. Positive and negative potential in the +10kBT/e to −10kBT/e range is shown in blue and red, respectively. (C) Details of the KLC1TPR-JIP1C-terminterface. The amino acids involved in the interactions are shown as stick models with oxygen atoms colored in red, nitrogen in blue and carbon in gold and magenta for KLC1TPR and JIP1C-term, respectively. Hydrogen bonds and salt bridges are represented by dotted grey and magenta lines, respectively. Residues tested for cellular validation (see panel D) are boxed. (D) Western blot analysis of coimmunoprecipitation assays showing the effect of selected KLC1TPR mutations on the interaction with JIP1. (E) Quantification of relative binding from two independent coimmunoprecipitation experiments. Error bars show SEM. All selected mutations reduce binding between ~65% and 90% compared to wild-type (WT) and are statistically significant to p<0.001. Statistical analysis was performed with the GraphPad Prism package using ordinary one-way ANOVA for multiple comparisons. (F) FP measurements show that the KLC2TPR S328,454N variant binds JIP1C-term with an affinity comparable to KLC1TPR with the S328N substitution playing a dominant role in restoring KLC1TPR-like affinity. Residues S328 and S454 of KLC2 correspond to residues N343 and N469 of KLC1TPR, respectively. Binding curves for KLC1TPR and KLC2TPR are also shown for comparison.
-
Figure 3—supplement figure 1—source data 1
Fluorescence polarization data for TAMRA-JIP1 amidated C-term binding to KLC1TPR.
- https://doi.org/10.7554/eLife.38362.013
-
Figure 3—source data 1
Quantification of relative KLC1TPR binding in three independent coimmunoprecipitation experiments as shown in panel E.
- https://doi.org/10.7554/eLife.38362.019
-
Figure 3—source data 2
Normalized fluorescence polarization and SEM for experiments presented in panel F.
- https://doi.org/10.7554/eLife.38362.020
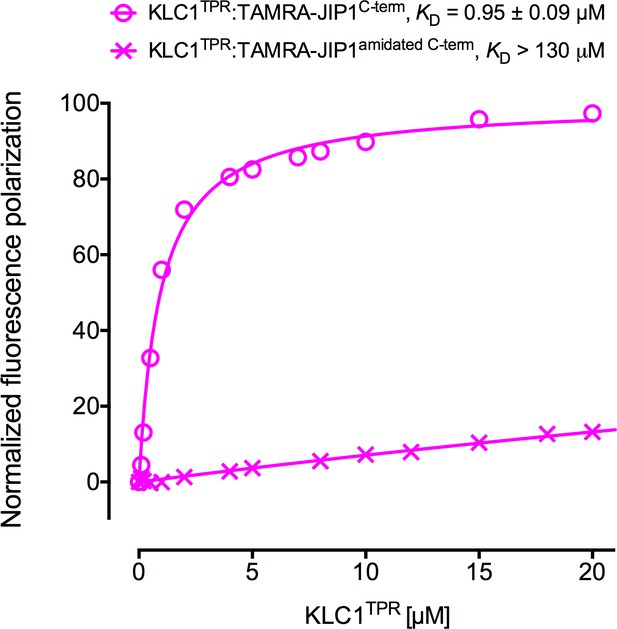
C-terminal amidation of JIP1C-term abrogates KLC1TPR binding.
FP measurements show that the loss of the C-terminal negative charge completely disrupts JIP1C-term binding to KLC1TPR. The binding curve of TAMRA-JIP1C-term to KLC1TPR is shown for comparison.
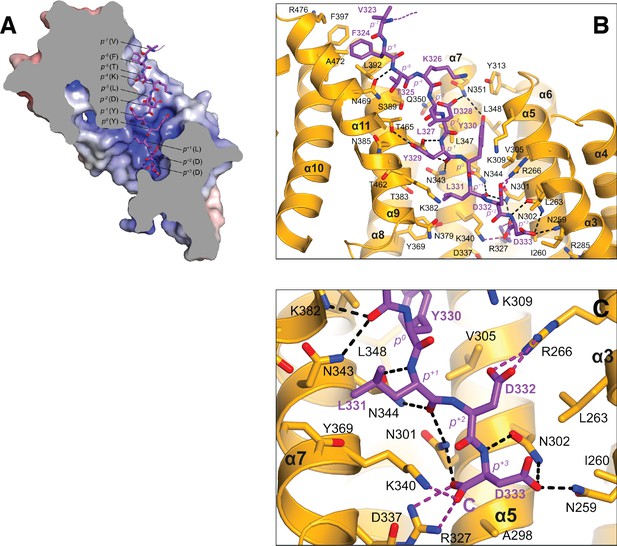
KLC1TPR-TorsinAC-term interface.
(A) Sliced-surface view of the KLC1TPR domain with bound TorsinAC-term. The molecular surface is colored according to its electrostatic potential. Positive and negative potential in the +10kBT/e to −10kBT/e range is shown in blue and red, respectively. (B) Details of the KLC1TPR-TorsinAC-term interface. Hydrogen bonds and salt bridges are represented by dotted black and magenta lines, respectively. (C) Close-up of the KLC1TPR-TorsinAC-term interface to highlight the interactions of the TorsinAC-term p+3 position that is not present in JIP1C-term. This view is slightly rotated compared to (B).
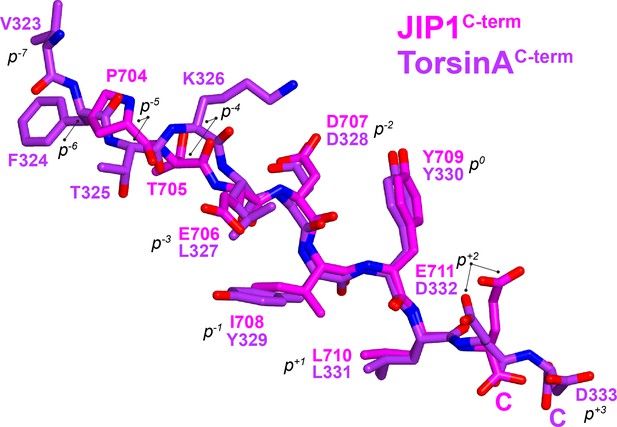
Superposition of KLC1TPR-bound JIP1C-term and TorsinAC-term peptides.
Carbon atoms belonging to JIP1C-term are shown in magenta while those of TorsinAC-term are in violet. Nitrogen and oxygen atoms are colored in dark blue and red, respectively. Peptide residues are labelled according to the sequence number and also using their common tyrosine as internal reference (position 0, p0). C-termini are highlighted with a letter C. Both peptides bind in an extended conformation with the (p−2-p+1) region displaying virtually identical main chain angles, while peripheral residues exhibit more variability.
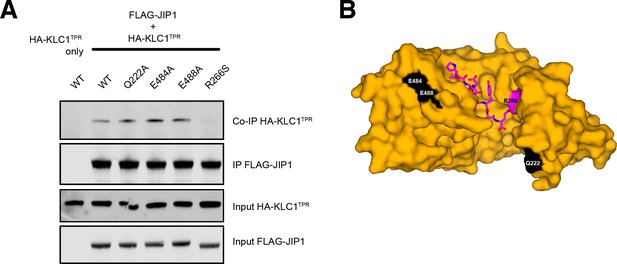
Control coimmunoprecipitation assays.
(A) Western blot analysis of coimmunoprecipitation assays showing the effect of selected KLC1TPR mutations on the interaction with JIP1. Residues Q222, E484, E488 are away from the interface and their substitution to an alanine does not decrease JIP1 binding. The R266S substitution at the interface is also shown for comparison. (B) Surface representation of JIP1C-term-bound KLC1TPR highlighting the residues in (A). R266 is shown in magenta. Q222, E484, E488 are highlighted in white. JIP1C-term is shown in magenta as sticks.
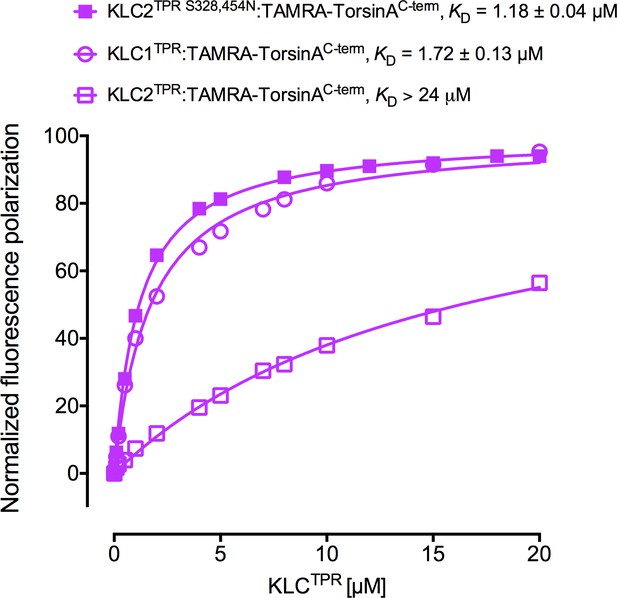
Fluorescence polarization measurements for the binding of TAMRA-TorsinAC-term to the KLC2TPR S328,454N variant.
The KLC2TPR S328,454N variant binds TorsinAC-term with an affinity comparable to KLC1TPR. Residues S328 and S454 of KLC2 correspond to residues N343 and N469 of KLC1TPR, respectively. Binding curves for KLC1TPR and KLC2TPR are also shown for comparison.
-
Figure 3—figure supplement 5—source data 1
Fluorescence polarization data for TAMRA-TorsinAC-term binding to KLC2TPR S328,454N .
- https://doi.org/10.7554/eLife.38362.018
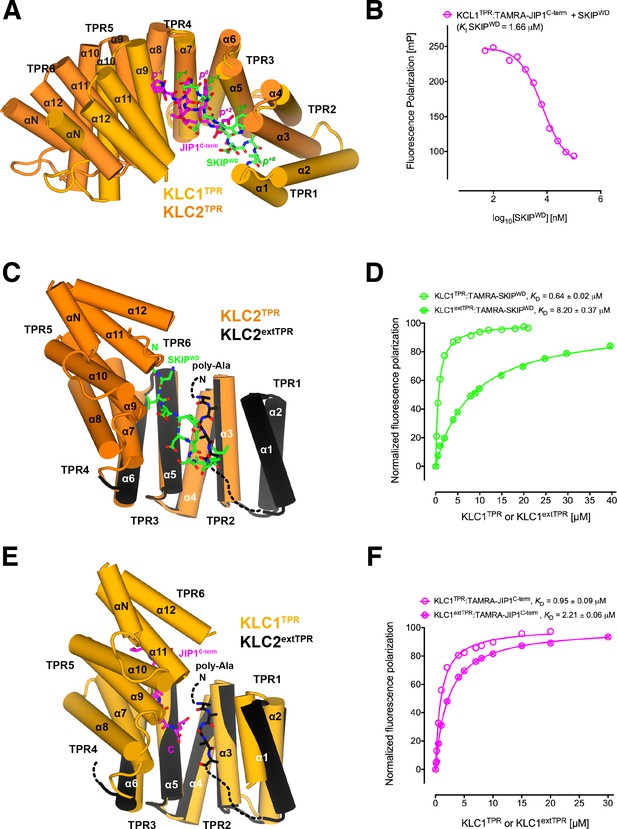
Y-acidic and W-acidic motifs bind at partly overlapping sites on KLCTPR and are differentially affected by the presence of the LFP-acidic intramolecular region.
(A) Superposition of the KLC1TPR-JIP1C-term and KLC2TPR-SKIPWD (Pernigo et al., 2013) complexes. JIP1C-term and SKIPWD are shown as stick representation and colored in magenta and green, respectively. Selected amino acid positions are labeled using the Y-acidic tyrosine and the W-acidic tryptophan as references (position 0, p0). KLC1TPR and KLC2TPR are colored in gold and orange, respectively. TPR1 is missing in the KLC2TPR-SKIPWD structure. The positions of the Y-acidic JIP1C-term and W-acidic SKIPWD peptides are shifted along the inner cavity of the super-helical KLCTPR architecture and binding at these alternative locations induces a different degree of conformational change in the receptor. (B) Titration of increasing amounts of unlabeled SKIPWD in the KLC1TPR:JIP1C-term complex shows a decreased FP signal that supports competitive displacement owing to their overlapping binding sites. (C) Superposition of the KLC2TPR-SKIPWD (Pernigo et al., 2013) and KLC2extTPR (Yip et al., 2016) structures (view is rotated by ∼90° around the x axis compared to A). For clarity, only the first three TPRs of KLC2extTPR (in black) that define the local environment of the LFP-acidic (also in black, modelled as poly-Ala) are shown. SKIPWD and LFP-acidic peptides bind at overlapping positions. (D) FP measurements show that the N-terminal extension of KLC1TPR domain that includes the LFP-acidic region (KLC1extTPR) inhibits the interaction with TAMRA-SKIPWD compared with KLC1TPR alone. (E) Superposition of the KLC1TPR-JIP1C-term and KLC2extTPR (Yip et al., 2016) structures (same orientation as C). As in (C) only the first three TPRs of KLC2extTPR are shown in black. JIP1C-term (in magenta) and LFP-acidic (in black) peptides bind at largely independent positions. (F) FP measurements show that the LFP-acidic N-terminal extension to KLC1TPR has only a modest impact on TAMRA-JIP1C-term binding (Yip et al., 2016). This panel is re-produced under the following license agreement: http://www.pnas.org/sites/default/files/advanced-pages/authorlicense.pdf. It is not available under CC-BY and is exempt from the CC-BY 4.0 license.
-
Figure 4—source data 1
Fluorescence polarization data for the competition of TAMRA-JIP1C-term with increasing amounts of unlabeled SKIPWD as shown in panel B.
- https://doi.org/10.7554/eLife.38362.023
-
Figure 4—source data 2
Normalized fluorescence polarization data and SEM for the experiments presented in panels E and F.
- https://doi.org/10.7554/eLife.38362.024
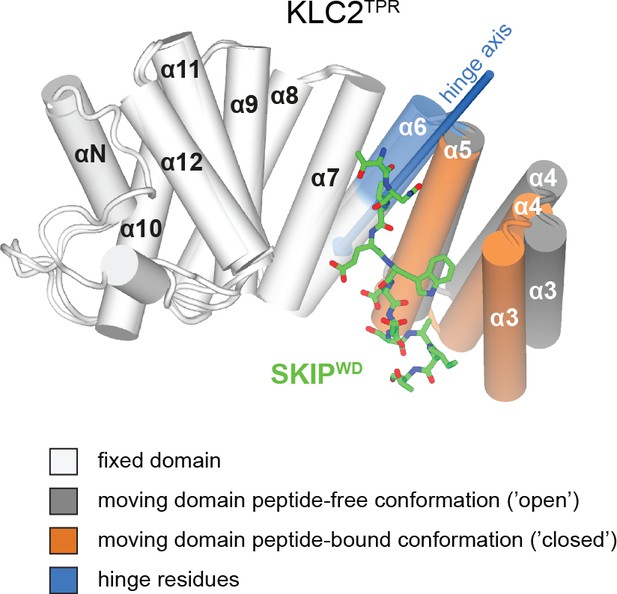
Conformational change in the KLC2TPR domain upon SKIPWD W-acidic peptide binding.
Superposition of ligand-free KLC2TPR (PDB code 3CEQ, chain B) and SKIPWD-bound KLC2TPR (PDB code 3ZFW, chain A) performed using the DynDom algorithm. The analysis reveals the presence of an N-terminal moving domain (residues 238 – 297, highlighted in orange and dark grey for KLC2TPR-SKIPWD and KLC2TPR, respectively) and a C-terminal fixed domain (residues 298 – 477, in light grey) while the (296 - 305) region represents bending residues (in blue). The conformational transition requires a rotation of ∼20° degrees around a hinge axis (blue arrow) that is approximately aligned with helix α6. TPR1 is not shown as it is not present in the X-ray structure of SKIPWD-bound KLC2TPR.
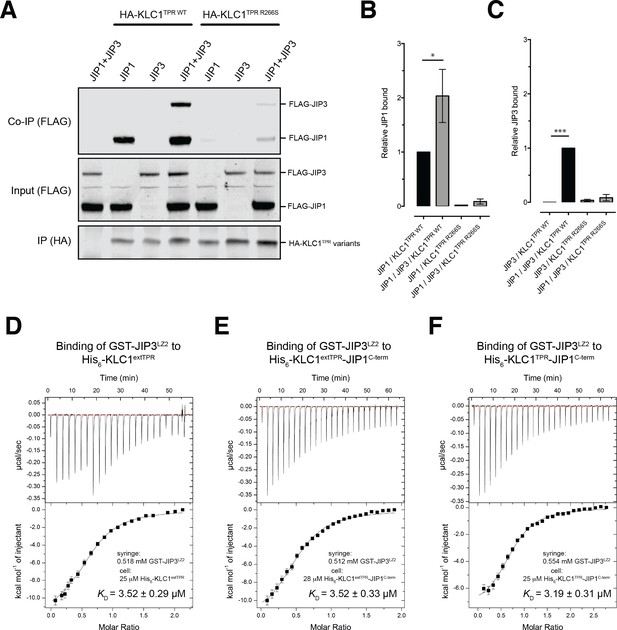
JIP1/JIP3 co-operativity.
(A) Western blot analysis of coimmunoprecipitation assays showing co-operative formation of the JIP1 and JIP3 complex. The effect of the KLC1TPR R266S mutations on the formation of the complex is also shown. (B, C) Quantification of relative JIP1 binding (B) and JIP3 binding (C) from three independent co-immunoprecipitation experiments (normalized to the sample indicated by a black bar). Error bars show SEM. Statistical analysis was performed with the GraphPad Prism package using ordinary one-way ANOVA for multiple comparisons, *p<0.05, ***p<0.001. (D, E, F) Binding of GST-JIP3LZ2 to His6-KLC1extTPR (D), His6-KLC1extTPR-JIP1C-term (E), His6-KLC1TPR-JIP1C-term (F) measured by ITC. It shows that JIP3LZ2 affinity for the TPR domain is not affected by either the LFP-acidic region or JIP1C-term loading. All experiments were carried out in 50 mM HEPES, 500 mM NaCl, 5 mM 2-mercaptoethanol at T = 20°C. In (D) the volume of the first six injections following the initial sacrificial one was 1.0 μl and the rest were 2.0 μl. In (E) and (F) all working injections were 1.5 μl. All injections were performed at 150 s time intervals.
-
Figure 5—source data 1
Quantification of relative JIP1 and JIP3 binding in three independent coimmunoprecipitation experiments as shown in panels B and C, respectively.
- https://doi.org/10.7554/eLife.38362.027
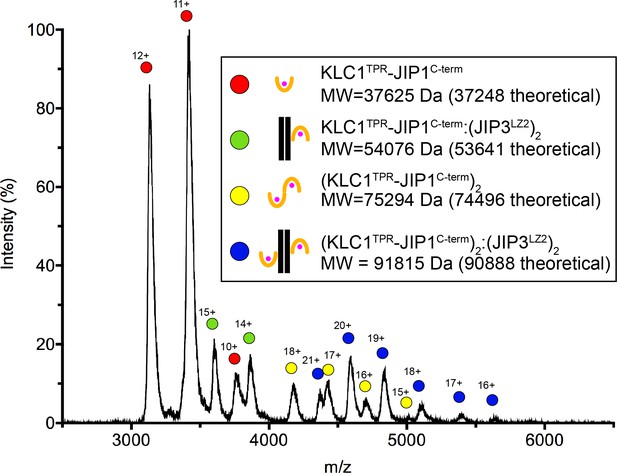
Native mass spectra of the untagged SEC-purified KLC1TPR-JIP1C-term:JIP3LZ2 complex.
Using an untagged SEC-purified KLC1TPR-JIP1C-term:JIP3LZ2 complex assembled using a slight molar excess (1.5-fold) of the JIP3LZ2 dimer, we find a mixture of KLC1TPR-JIP1C-term monomers and dimers both in isolation and bound to JIP3LZ2 indicating that up to two KLC1TPR-JIP1C-term molecules can bind to the coiled-coil. The illustrations indicate the various species observed and alongside their corresponding molecular weights (MW).
Videos
KLC1TPR conformational transition upon TorsinAC-term binding.
https://doi.org/10.7554/eLife.38362.010Tables
X-ray data collection and refinement statistics.
https://doi.org/10.7554/eLife.38362.007| Data collection | ||
|---|---|---|
| Data set | KLC1TPR-JIP1C-term | KLC1TPR-TorsinAC-term |
| Beam line | P14 (Petra) | I04 (DLS) |
| Wavelength (Å) | 0.9763 | 0.9795 |
| Resolution range* (Å) | 45.18–2.70 (2.83–2.70) | 52.50–2.29 (2.83–2.70) |
| Space group | C2 | C2 |
| Cell dimensions (a, b, c) (Å) (α, β, γ) (°) | 107.47, 90.36, 51.70 90, 99.80, 90 | 106.04, 89.68, 50.99 90, 98.00, 90 |
| Unique reflections* | 13229 (1747) | 21275 (1021) |
| Overall redundancy* | 3.8 (3.9) | 4.7 (3.2) |
| Completeness* (%) | 98.6 (98.5) | 99.6 (93.9) |
| Rmerge* (%) | 5.1 (117.6) | 8.0 (83.2) |
| Rp.i.m. (I)* (%) | 4.7 (106.7) | 4.0 (52.0) |
| ⟨I/σ(I)⟩* | 11.9 (1.0) | 9.0 (1.1) |
| CC(1/2)* (%) | 99.9 (58.4) | 99.7 (74.1) |
| Refinement | ||
| PDB code | 6FUZ | 6FV0 |
| Rfactor (%) | 23.3 | 20.8 |
| Rfree (%) | 25.6 | 24.5 |
| # non-H atoms protein water heterogen | 3001 13 6 (GOL) | 3039 90 7 (PEG) |
| Average B value (Å2) | 113.04 | 65.81 |
| rms bond lengths (Å) | 0.009 | 0.007 |
| rms bond angles (°) | 1.08 | 0.93 |
-
*Numbers in parentheses refer to the highest resolution bin.
ITC parameters.
https://doi.org/10.7554/eLife.38362.028| GST-JIP3LZ2 interactions with | N | KD (μM) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| His6-KLC1extTPR | 0.59 ± 0.01 | 3.52 ± 0.29 | −13.19 ± 0.38 | −5.86 | −7.33 |
| His6-KLC1extTPR-JIP1C-term | 0.52 ± 0.01 | 3.52 ± 0.33 | −12.69 ± 0.45 | −5.39 | −7.30 |
| His6-KLC1TPR-JIP1C-term | 0.72 ± 0.02 | 3.19 ± 0.31 | −7.69 ± 0.24 | −0.32 | −7.37 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38362.029





