Importance of miRNA stability and alternative primary miRNA isoforms in gene regulation during Drosophila development
Figures
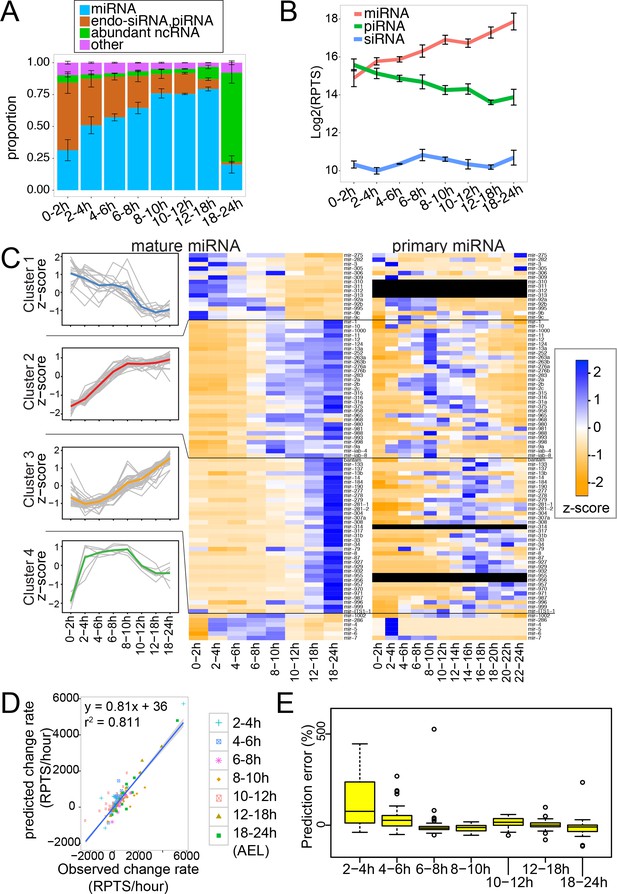
Mature and pri-miRNA profiles revealed by RNA-seq analysis.
(A) Composition of small RNA libraries. Small RNA reads were grouped into four categories, and their fractions in the library were plotted. The average and standard error of mean of three replicate libraries are shown. (B) Changes in the global levels of small RNA classes. The averages of total read counts of miRNAs (red), piRNAs (green) and siRNAs (blue) were plotted. Error bars indicate standard error of mean. Small RNA read counts were normalized against the spike-in count of each library and expressed as reads per thousand spike-in reads (RPTS). (C) miRNA genes were grouped into four clusters by k-means clustering (k = 4) and expression z-scores of individual miRNAs are shown in the left panel (gray lines). The average z-score for each cluster is shown as a colored line. Expression profiles of mature miRNAs (middle) or pri-miRNAs (right) in staged fly embryos are shown in the heatmap. The heatmap was color-coded according to the z-score calculated across the 8 or 12 time-windows for each miRNA gene. The pri-miRNA level was determined as the read density in the 100nt window immediately upstream of the miRNA hairpin. (D) The mature miRNA change rates predicted by multiple regression analysis were plotted against observed values. (E) Error rates estimation for each time window. Error rates were determined by the following formula. Error rates = (predicted - observed miRNA change rate) * (window size) * 100 / (mature miRNA level). Distributions of prediction error rates were plotted. Four outliers in the 2–4 hr time window (1402.5, 972.8, 954.3 and 739.9%) are not shown. Genes with at least 10 RPTS in the time window were used for this analysis.
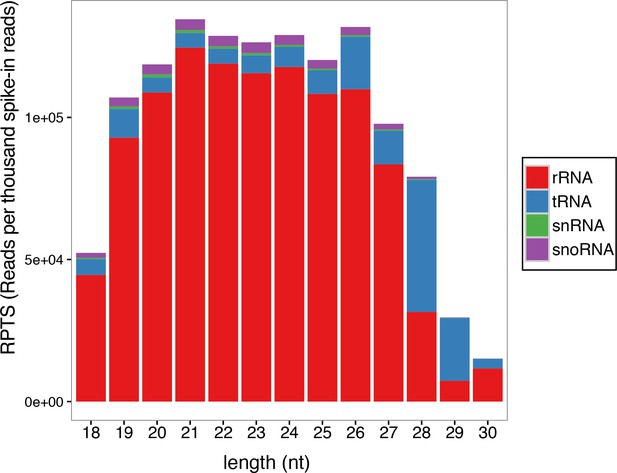
Size distribution of small RNA reads mapping to abundant ncRNAs.
Small RNA reads belonging to the ‘abundant ncRNA’ category in the small RNA libraries made from 18 to 24 hr embryos were divided into four subgroups based on their origins (rRNA, tRNA, snRNA and snoRNA). The average of normalized read counts for each subgroup in the triplicate libraries was plotted for each length of small RNAs. Read counts were normalized by the number of external spike-ins, and expressed as reads per thousand spike-in reads (RPTS).
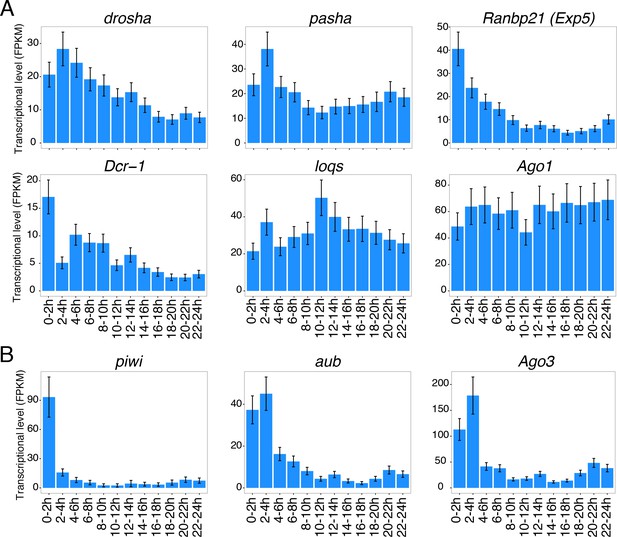
mRNA levels of miRNA/piRNA processing factors.
mRNA expression levels for (A) miRNA processing factors and (B) piRNA processing factors during fly embryogenesis were estimated using total RNA-seq data. Averages and standard error of mean of FPKM (Fragment Per Kilobase of transcripts per Million mapped reads) values are shown.
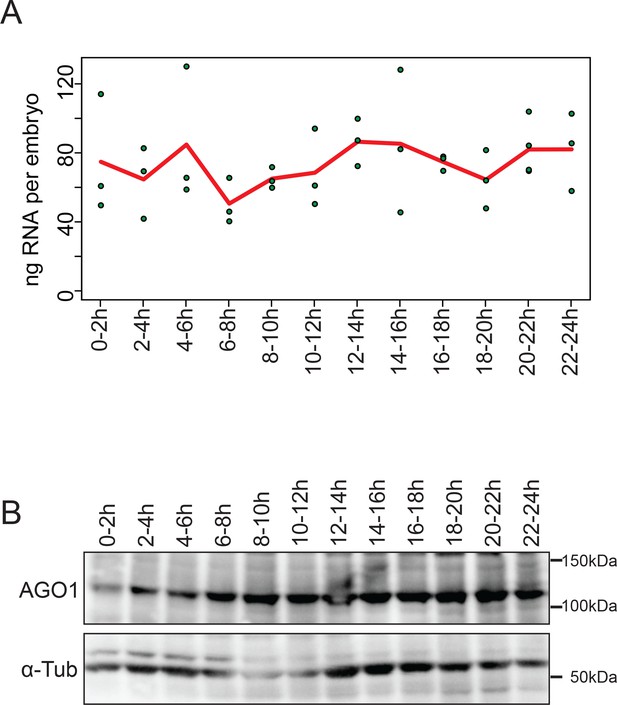
Relative levels of total RNA and AGO1 protein per embryo in developing embryos.
Levels of (A) total RNA and (B) AGO1 protein. Staged embryos from the twelve 2 hr windows were used. (A) The RNA levels were examined by UV absorbance of total RNA isolated from 100 embryos. The experiments were done in tripricate, and the means and individual values were shown as the red line and green circles, respectively. (B) Western blotting results using anti-AGO1 or anti-ß-tubulin antibody. A total protein samples from 20 embryos was used in each lane.
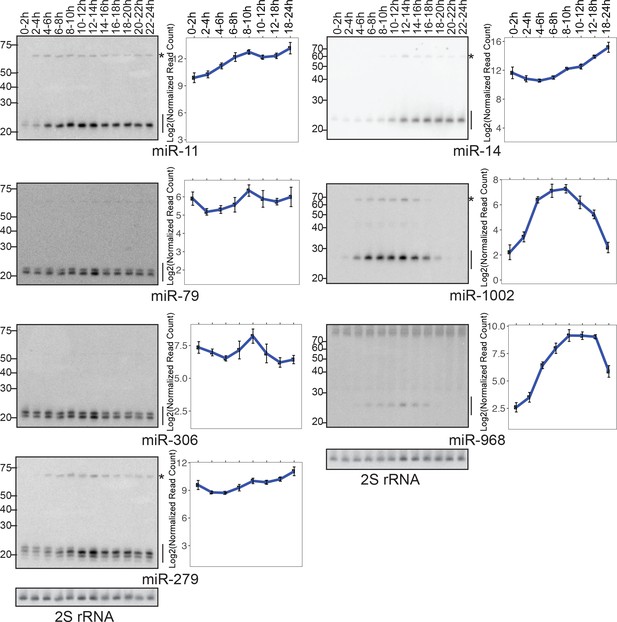
Verification of expression data by Northern blotting.
(Left panels) RNA samples from staged embryos were resolved by 15% denaturing PAGE and probed for the mature species of the indicated miRNA gene. Mature miRNA and precursor species are indicated by lines and asterisks respectively. (Right panels) Mature miRNA quantification using small RNA library data. Average values of triplicates are shown. Error bars indicate the standard error of mean.
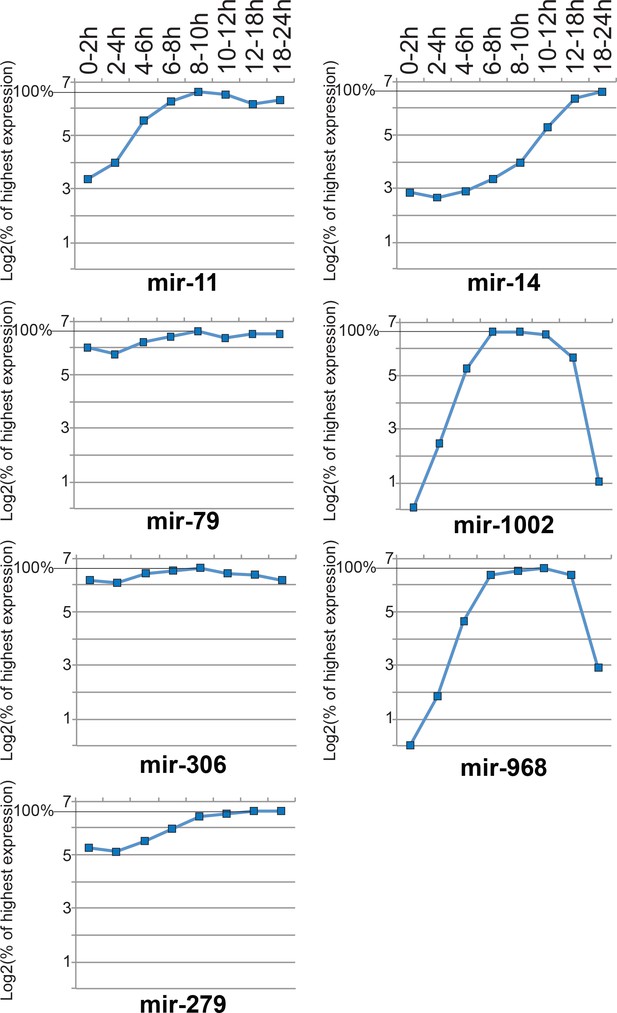
Quantification of Figure 2 Northern blotting results.
Mature miRNA signals were quantified and normalized by the 2S rRNA signal in the corresponding lane. The expression values were further normalized by the signal from the time window where the highest expression level of the miRNA was seen, and expressed as a percentage. The y-axes show log2 of the normalized expression values.
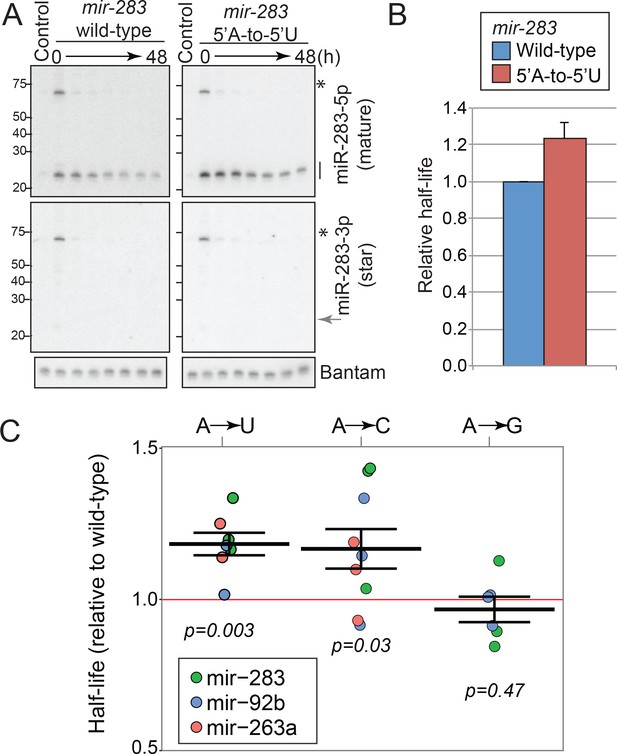
Effects of the 5’ nucleotide on miRNA stability.
(A) Experimental validation of the role for miRNA 5’ nucleotides in mature miRNA stability. A genomic DNA fragment containing mir-283-cluster locus was cloned in a CuSO4-inducible plasmid (mir-283 wild-type). The 5’-nucleotide mutation was introduced by site-directed mutagenesis to generate the mutant plasmid (mir-283 A-to-U) and the plasmids were used for transfection of S2-R+ cells. Transfected cells were cultured for 24 hr in the presence of CuSO4 to induce the expression, and the expression was terminated by replacing with medium containing the CuSO4 chelator (time 0). The time intervals after the withdrawal of the inducer are: 0, 6, 12, 24, 30, 36 and 48 hr. For control lanes, cells were transfected with the same plasmid but CuSO4 induction was omitted. RNA samples were prepared for Northern blotting analysis at the indicated time points to monitor the reduction rates of mature miRNA species. Asterisks and lines indicate the positions of precursor and mature miR-283 signals, respectively. A representative figure of three attempts is shown. The miR-283 star strand was present at very low levels compared to the mature strand, confirming that the miRNA stability measurements reflect the half-lives of mature miRNAs that are already loaded to the Argonaute complex. Endogenous bantam was detected and used as a loading control. (B) Quantification of the triplicates of miR-283 mutant analysis shown in (A). The relative miRNA half-life for miR-283 (A–to–U) was normalized by that from the corresponding wild-type result in the same replicate. The average and the standard deviation are shown. The miR-283 A-to-U mutant exhibited a slightly extended half-life (p=0.018, one-tailed paired t-test, N = 3). (C) Mature miRNA stability of mutant miRNAs. 5’ ends of the mature strands were mutated to indicated nucleotides and the time course experiment was done using S2-R+ cells expressing the wild-type or mutant miRNAs under the CuSO4-inducible promoter, similar to Figure 6B and C. The relative half-life of the mutant miRNA was determined for each replicate, and individual values (dots) and means ±standard errors (lines) are shown. The colors of dots indicate the miRNA backbone used for mutagenesis. Student’s t-test p-values are shown in the chart. Representative images can be found in Figure 6—figure supplement 2. Raw data can be found in Figure 3C—source data 1.
-
Figure 3—source data 1
Raw data for Figure 3C.
The normalized half-lives for individual replicates that were used for drawing the chart are shown.
- https://doi.org/10.7554/eLife.38389.012
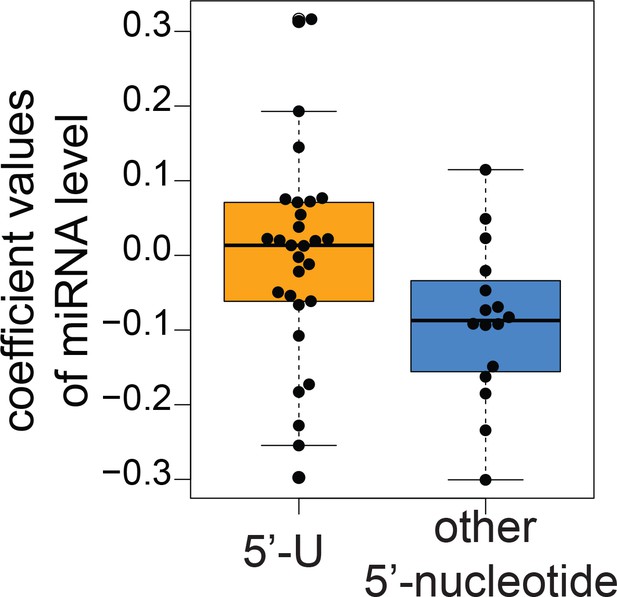
Distributions of estimated relative half-lives for 5’-U miRNAs and miRNAs with other 5’ nucleotides.
Distribution of coefficient values associated with the mature miRNA abundance determined by the multiple regression analysis shown in Supplementary file 4. We expect the coefficients to reflect the degradation rates therefore negative values. Note that there were many positive coefficients, suggesting that this analysis was not very accurate. Distributions of coefficients for 5’-U miRNAs (orange) and non-5’-U miRNAs (5’-nucleotide = G, C or A; blue) are shown.
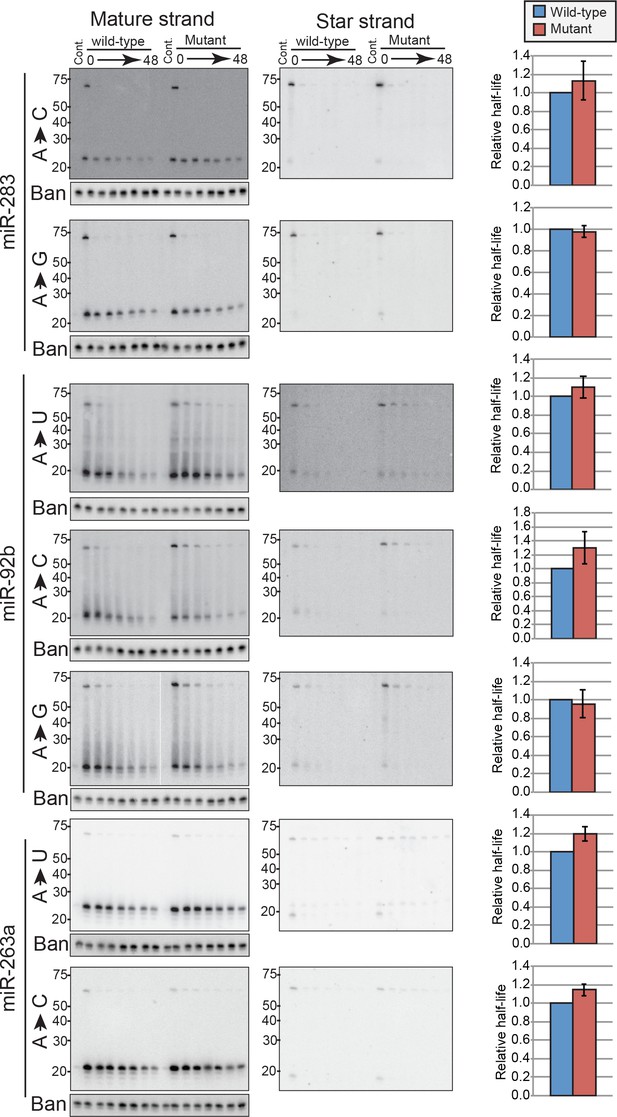
Mutagenesis of 5’ nucleotides in mir-283, mir-92b and mir-263a backbones.
Representative images of 5’ nucleotide mutant experiments summarized in Figure 6D are shown. The experimental setting and time intervals are same as Figure 6B. The bar charts show relative half-lives of respective 5’ mutants, in the same format as Figure 6B.
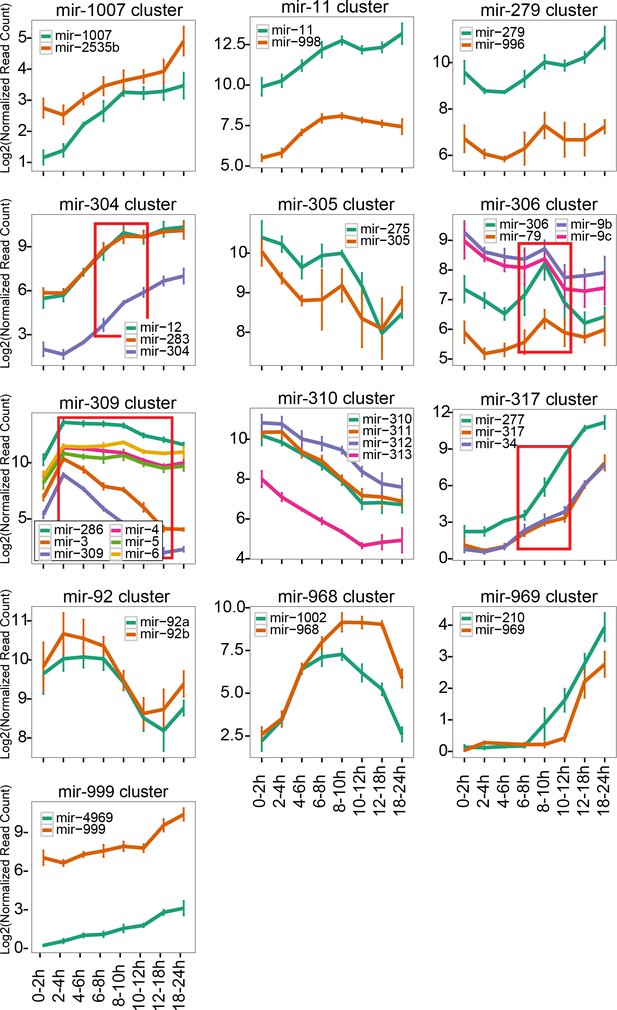
Expression changes of clustered miRNAs.
Normalized read counts of individual miRNA clusters were plotted. The lines and error bars indicate the averages and standard error of mean respectively. The time windows showing significant expression changes are indicated by red rectangles (ANOVA p<0.01, N = 3, p-values are shown in Supplementary file 5).
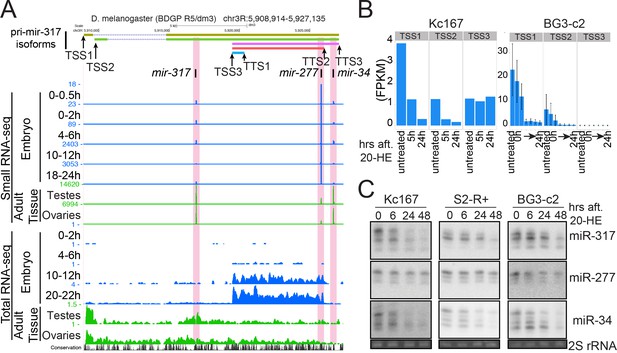
Distinct miRNA subsets are produced from the mir-317 cluster by alternative pri-miRNA isoforms.
(A) UCSC genome browser snapshot of the mir-317 cluster locus. Small RNA-seq (upper) and total RNA-seq (lower) data are shown. In embryos (blue) miR-277 expression is higher than the other two miRNAs, and even higher in late embryos, which is consistent with the high expression of pri-miRNA isoform starting from TSS3 in embryos. In testes and ovaries, long isoforms starting from TSS1 and 2 are dominant, and the miR-277 level is lower than miR-317 and miR-34. In all tissues and embryonic stages, the ratios between miR-317 and miR-34 remain similar. (B) Reanalysis of published total RNA-seq data from cultured cells treated with ecdysone. The sum of FPKMs of the isoforms sharing the same TSS was plotted. (C) Northern blotting analysis of mature miR-317,–277 and −34 levels after 20-HE (hydroxyecdysone) addition in Kc167, S2-R+ and BG3-c2 cells. Cells were treated with 20-HE for the indicated time and total RNA was separated on a 15% denaturing acrylamide gel. miR-317 and miR-34 were decreased after 20-HE addition in Kc167 and S2-R+ cells, while the decrease of miR-277 was much weaker in these cell lines. In contrast, miR-277 was also decreased in BG3-c2 cells, in which the level of the mir-277 specific short isoform was very low even in the absence of 20-HE (Panel B, TSS3). See Figure 5—figure supplement 3for the quantified results of tripricates.
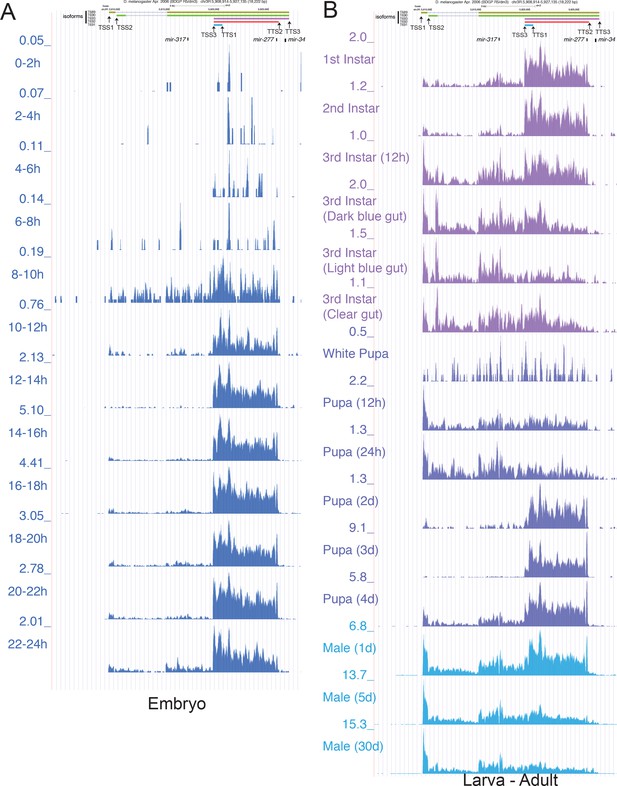
UCSC Genome Browser screen shot of the mir-317 cluster locus with total RNA-seq tracks.
(A) Total RNA-seq data from staged embryos show that the short pri-mir-317 isoform starting from TSS3 is the dominant isoform in embryos with an expression peak at 14-18hAEL. (B) Total RNA-seq data from post-embryonic stages show dynamic changes in pri-mir-317 isoform selection.
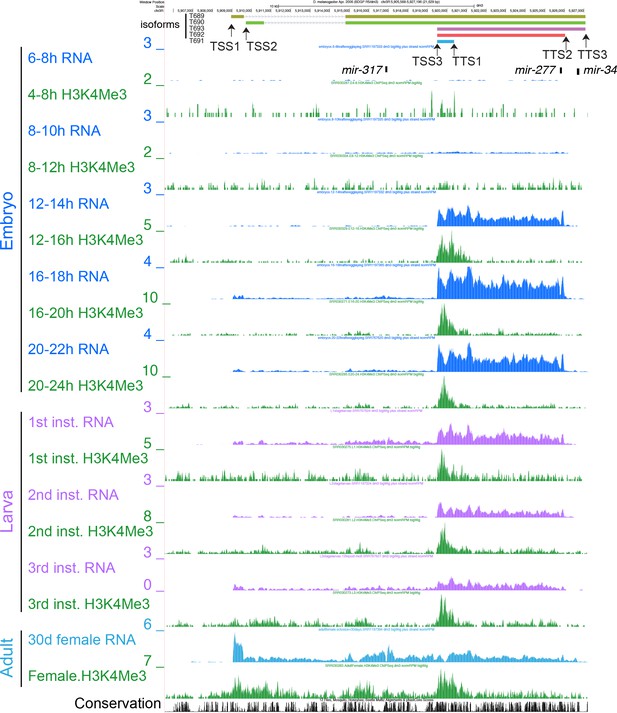
UCSC Genome Browser screenshot of the mir-317 cluster locus with total RNA-seq and Histone H3K4me3-ChIP tracks.
Total RNA-seq tracks are color coded by the developmental stage (Blue: embryo, Purple: larva, Light blue: adult). Green tracks are ChIP-seq tracks for H3K4me3, which is enriched at active chromatin regions (modENCODE Consortium et al., 2010). H3K4me3 peaks were seen near the TSS3 in late embryos, and additional H3K4me3 peaks appeared near TSS1/2 sites in adults, supporting the alternative usage of the TSSs identified by the RNA-seq analysis.
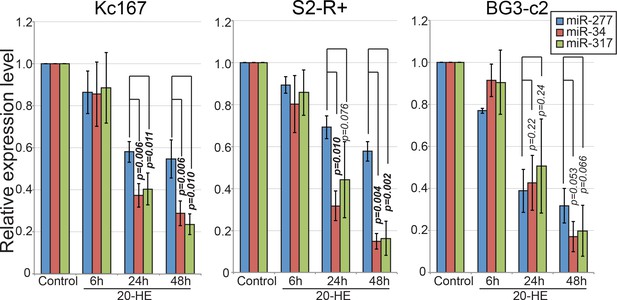
Quantification of the Northern blotting results shown in Figure 4C and their biological replicates.
Kc167, S2-R+ and BG3-c2 cells were treated with 20-HE for the indicated time and total RNA was separated on a 15% denaturing acrylamide gel. The mature signals were quantified and normalized for 2S rRNA signals. The normalized values from three replicates were plotted (Average ± standard deviation). Student’s t-test p-values are shown. P-values less than 0.05 are shown in bold letters. Raw data can be found in Figure 5—figure supplement 3—source data 1.
-
Figure 5—figure supplement 3—source data 1
Raw data for Figure 5—figure supplement 3—source data 1.
Individual values for the replicates (Sheet 1: Raw value), and averages and standard deviations (Sheet 2: Summary) are reported. For the charts, values on the summary sheet were used. T-test p-values are reported on the raw value sheet.
- https://doi.org/10.7554/eLife.38389.018
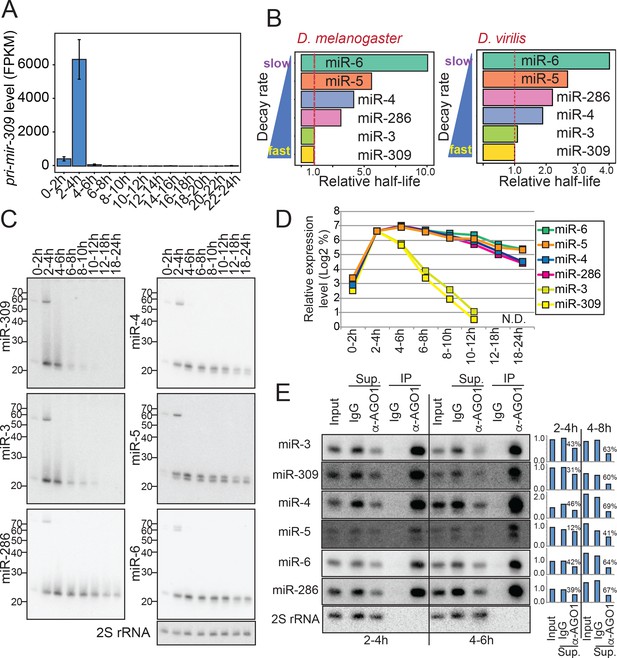
Differential mature miRNA half-lives for the mir-309 cluster genes.
(A) Levels of pri-mir-309 during embryogenesis. Transcript levels were quantified using the total RNA-seq libraries and normalized by the FPKM (Fragment Per Kilobase of transcripts per Million mapped reads) method. Pri-mir-309 is transiently expressed in 2–4 hr embryos. Error bars indicate the 95% confidence intervals. (B) Evolutionary conservation of the differential miRNA half-lives of mir-309 cluster miRNAs. Estimated relative half-lives in D. melanogaster (this study) and D. virilis based on a published dataset (Ninova et al., 2014) are shown. The values are normalized by the half-life of miR-309. Note that D. virilis libraries did not include the spike-in oligos, and the TMM (Trimmed Mean of M-values) normalization was used (Robinson and Oshlack, 2010). The D. melanogaster data were normalized by the spike-in counts. (C) Northern blotting analysis was performed with the RNA samples extracted from embryos in the indicated time windows and using the probes detecting the indicated mature miRNA species. (D) Quantification of panel (C). The expression value of each miRNA in 0–2 hr sample was set as 100% and relative levels in each time window was calculated. The Y-axis shows log2 of % expression values. (E) Mature miRNA species from the mir-309 cluster are efficiently loaded in the AGO1 complex. Lysates were prepared from 2-4 hr and 4–6 hr old embryos, and the AGO1 complex was precipitated using anti-AGO1 antibody. Rabbit IgG was used as a negative control. Efficient precipitation was confirmed by the enrichment of mature miRNA species in the AGO1-IP lane, and the depletion of the mature miRNA species in the AGO1-IP supernatant (Sup.) lane. The mature miRNA signals were quantified and normalized by the corresponding 2S rRNA signals in input and supernatant lanes. The input signal intensity at the 2–4 hr time window was used for further normalization for each miRNA species. Normalized values were plotted in the bar charts. The percentages in the charts indicate the degrees of mature miRNA depletion in the supernatant after AGO1-IP compared to the IgG control supernatant. We did not observe a correlation between miRNA stability and the degree of depletion by AGO1-IP, excluding the possibility that the differential half-lives of mature miRNAs from this cluster was caused by differential loading efficiencies of the miRNAs.
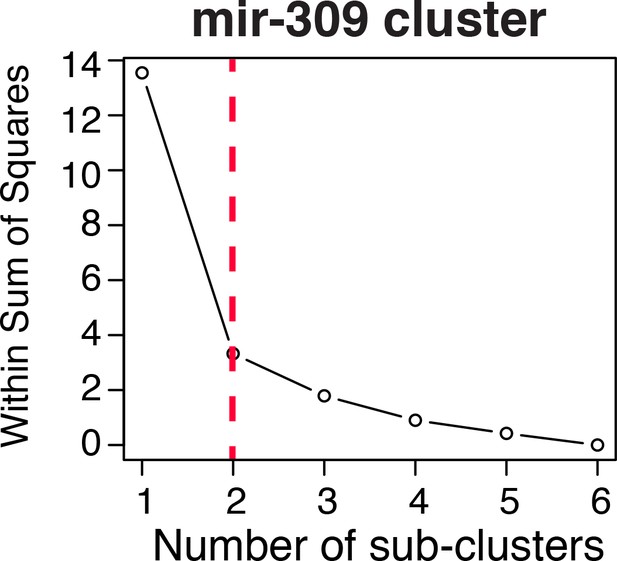
Elbow plot analysis for mir-309 cluster.
The steep drop in the within sum of squares between 1 and 2 sub-clusters suggests that the patterns of miRNA expression changes for the mir-309 cluster members can be grouped into two sub-clusters.
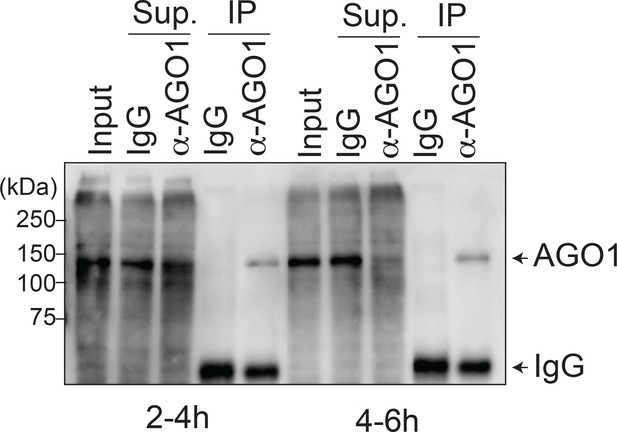
Detection of AGO1 protein in immuno-precipitates used for Figure 5 (E).
Western blotting analysis showed that AGO1 protein was successfully immuno-precipitated from lysates prepared from embryos collected in the 2–4 hr or 4–6 hr time windows.
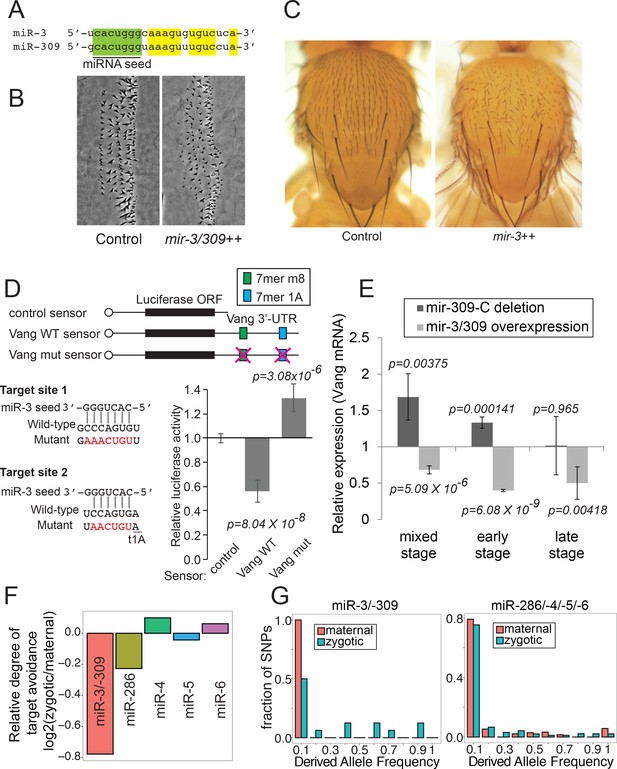
Biological activity of miR-3/–309.
(A) Sequences of mature miR-3 and miR-309. Note that their seed sequences are identical. (B) Embryonic denticles in control and embryos overexpressing miR-3/–309 (da-gal4 - > UAS-mir-3/–309). Misorientation of denticles was observed in embryos overexpressing miR-3/–309. (C) Misorientation of adult sensory bristles on the adult notum by miR-3 overexpression (eq-gal4 - > UAS-mir-3). (D) Luciferase sensor assays. S2-R+ cells were transfected with plasmids carrying a luciferase sensor containing the Vang 3’ UTR sequence along with a plasmid to overexpress miR-3 or a negative control empty vector. The averages and standard deviations of normalized Luciferase activity are shown (N = 8). The p-values were calculated by t-test. Vang 3’ UTR contains two predicted miR-3 family target sites. Removal of these predicted target sites abolished down-regulation of the Vang sensor by overexpressed miR-3. Raw data can be found in Figure 7D-source data 1. (E) Expression levels of Vang mRNA determined by quantitative RT-PCR. Values were normalized against RpL32. Embryos were collected at three time windows (mixed: 0–24 hr, early:0–12 hr or late:12–24 hr) using control, a deletion mutant lacking the mir-309 cluster (mir-309-C) miRNAs (dark gray) and a transgenic line overexpressing miR-3/–309 (light gray). The average relative expression ±standard deviation and p-values are shown (t-test). Vang mRNA is up-regulated in the deletion mutant whereas down-regulated in embryos overexpressing miR-3/–309 in the mixed stage samples. Note that derepression of Vang was not significant in null mutant embryos in the late stage consistent with the low level of miR-3/–309 in late embryos due to quick degradation. (F) To calculate relative target avoidance, the fractions of polymorphic target sites with target allele frequencies < 0.1 were computed for maternal and zygotic mRNAs. The ratio of fractions (Fractionzygotic/Fractionmaternal) was defined as relative degree of target avoidance. miR-3/–309 polymorphic target sites found in zygotically expressed mRNAs showed a weaker degree of target avoidance compared to those in maternal mRNAs, while much smaller differences were seen in the degree of target avoidance for target sites of other members of mir-309 cluster miRNAs. (G) Distributions of derived target allele frequencies (DAF) on maternal and zygotic genes for miR-3/–309 targets (left) or miR-286/–4/−5/–6 (right) were plotted. The zygotic DAF distribution for miR-3/–309 targets was shifted to the right compared to that of maternal genes (D = 0.56, p=0.046, Nmaternal = 7, Nzygotic = 16; one-tailed Kolmogorov-Smirnov test). The difference was less significant for the miR-286/–4/−5/–6 target set (D = 0.18, p=0.078, Nmaternal = 52, Nzygotic = 131). Raw data for (F) and (G) are reported in Supplementary file 6.
-
Figure 7—source data 1
Normalized Renilla (sensor)/firefly (control) luciferase activity ratios that were normalized to the psiCHECK empty vector value for each of the pDsRed and pDsRed-miR-3 groups are shown in the first 13 rows.
Rows 3–10 show values of individual replicates, and averages and standard deviations of the eight replicates are shown in rows 12 and 13. The values were further normalized by the values of corresponding sensor values in the pDsRed group, and the averages and standard deviations are shown in rows 16–19. The values in rows 16–19 were used for the chart.
- https://doi.org/10.7554/eLife.38389.026
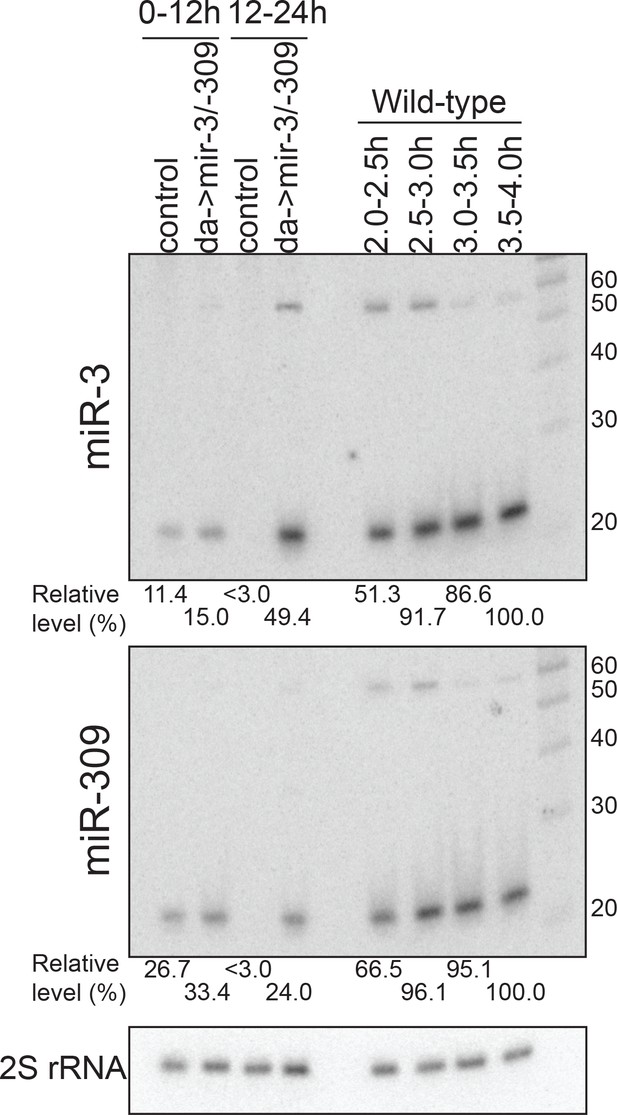
Levels of miR-3/–309 overexpression.
RNA samples used for the RT-PCR assay (Figure 7E) or RNA samples from staged wild-type embryos (30 min windows in 2–4 hr) were analyzed by Northern blotting. 10 ug total RNA was loaded in each lane and the membrane was probed for miR-3, miR-309 and 2S rRNA. Signals were quantified and the miR-3 and miR-309 signals were normalized by the 2S rRNA signal, and further normalized by the values in the 3.5–4.0 hr wild-type embryo sample. The results indicated that the level of overexpressed miR-3 and miR-309 did not exceed the level of endogenous miR-3 and miR-309 at the highest peaks of these miRNAs.
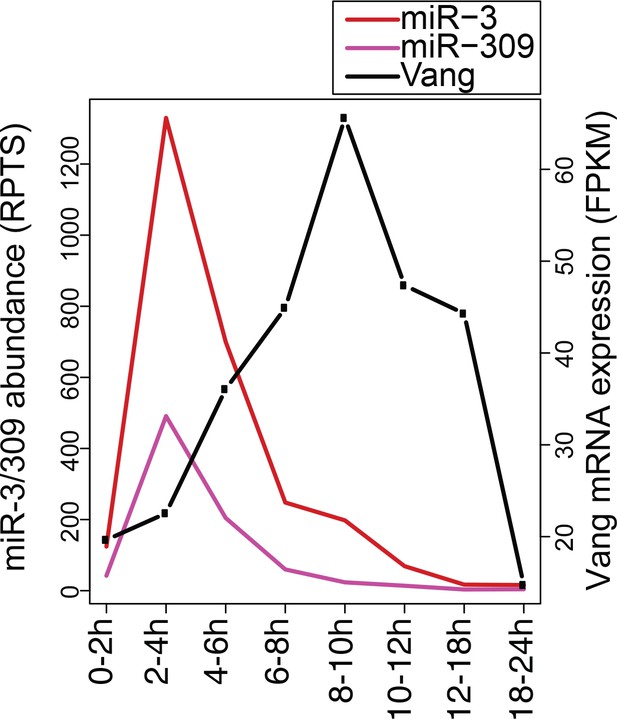
Expression levels of miR-3, miR-309 and Vang mRNA.
Relative expression levels of miR-3, miR-309 and Vang mRNA were determined using small RNA or total RNA-seq libraries and RPTS (for miRNAs) and FPKM (for mRNA) values were plotted. The increase of Vang mRNA was seen during the 2–10 hr time window, where the mature miR-3 and miR-309 levels were quickly decreasing.
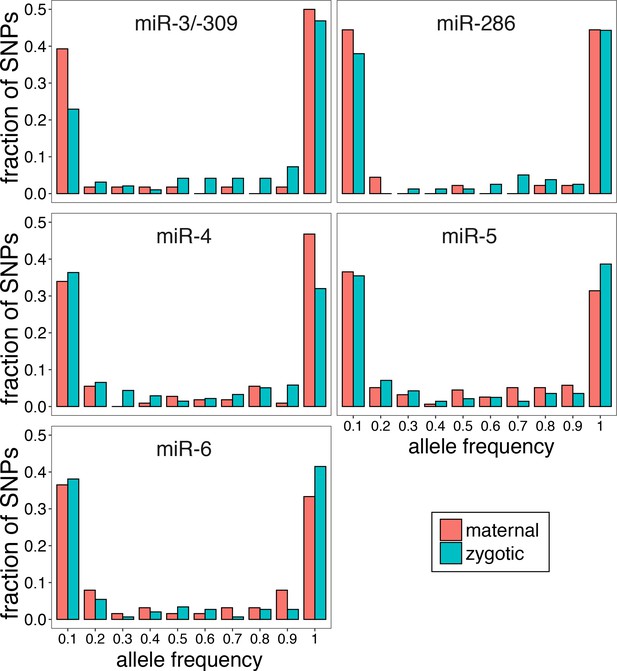
Allele frequency distribution of polymorphic miRNA seed target sites.
Polymorphic seed complementary sites for the mir-309 cluster miRNAs occurring in mRNA 3’UTRs were computationally identified and their allele frequencies were calculated using the DGRP data (Mackay et al., 2012). Protein coding genes were grouped into maternal and zygotic categories based on expression patterns according to the Paris definition (Paris et al., 2015). The fraction of polymorphic sites in maternally (red) or zygotically (green) expressed genes in each 0.1 frequency bin was plotted. The figure format follows the convention defined by a previous study (Marco, 2015), where the 0.9–1.0 bin contains sites with >90% of the population carrying the major allele that corresponds to the seed complementary sequence. For Figure 7F, the ratio of the 0–0.1 bars for zygotic and maternal mRNAs for each miRNA seed species was used to estimate relative degree of target avoidance.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | Eq-Gal4 | Bloomington Drosophila Stock Center | BDSC:43659 | FlyBase symbol: P{GAL4-Hsp70.PB}l(3)Eq1Eq1 |
| Genetic reagent (D. melanogaster) | Da-Gal4 | Bloomington Drosophila Stock Center | BDSC:55851 | FlyBase symbol: P{GAL4-da.G32}UH1 |
| Genetic reagent (D. melanogaster) | UAS-mir-3 | PMID: 22745315 | ||
| Genetic reagent (D. melanogaster) | UAS-mir-3/–309 | PMID: 26138755 | ||
| Genetic reagent (D. melanogaster) | mir-309-C delta 1 | PMID: 18394895 | ||
| Cell line (D. melanogaster) | S2-R+ | PMID: 9822716 | DGRC Cat# 150; RRID:CVCL_Z831 | S2-R + cells maintained in the Lai and Okamura labs |
| Cell line (D. melanogaster) | Kc167 | Drosophila Genomics Resource Center | DGRC Cat# 1; RRID:CVCL_Z834 | |
| Cell line (D. melanogaster) | BG3-c2 | Drosophila Genomics Resource Center | DGRC Cat# 6;, RRID:CVCL_Z728 | |
| Antibody | Anti-AGO1 (rabbit polyclonal) | Abcam | Abcam Cat# ab5070; RRID:AB_2277644 | 1:1000 in TBST |
| Antibody | Anti-alpha-tubulin (mouse monoclonal clone DM1A) | Sigma | Sigma-Aldrich Cat# T9026; RRID:AB_477593 | 1:1000 in TBST |
| Recombinant DNA reagent | psiCHECK2 (modified MCS) | PMID:17599402 | SalI-SacI-NotI-XbaI-SalI-EcoRI- EcoRV-XhoI-SpeI sites were inserted in the SalI/NotI sites of psiCHECK2 (Promega) | |
| Recombinant DNA reagent | psiCHECK-Vang wild-type | This study | Vang 3'UTR was amplified from genomc DNA using NotI_Vang_2128 and XhoI _Vang_3492 primers | |
| Recombinant DNA reagent | psiCHECK-Vang mutant | This study | The two mir-3 target sites were mutated by overlap PCR using Vang_t1_mut_F, Vang_t1 _mut_R, Vang_t2_mut_F and Vang_t2_mut_R and the cloning primers used for cloning of the wild-type sensor | |
| Recombinant DNA reagent | pUAST-DsRed-mir-3 | PMID: 22745315 | ||
| Recombinant DNA reagent | pUAST-DsRed | PMID: 12679032 | ||
| Recombinant DNA reagent | pRmHa-mir-283-C | This study | Genomic DNA fragment amplified by EcoRI_dme_mir283c_F and SalI_dme_mir283c_R inserted to pRmHa3 (DGRC: 1145). | |
| Recombinant DNA reagent | pRmHa-mir-283 A-to-U | This study | Site directed mutagenesis of pRmHa-mir-283-C using dme_mir283_5pAtoU_F and dme_mir283_5pAtoU_R | |
| Recombinant DNA reagent | pRmHa-mir-283 A-to-G | This study | Site directed mutagenesis of pRmHa-mir-283-C using dme_mir283_5pAtoG_F and dme_mir283_5pAtoG_R | |
| Recombinant DNA reagent | pRmHa-mir-283 A-to-C | This study | Site directed mutagenesis of pRmHa-mir-283-C using dme_mir283_5pAtoC_F and dme_mir283_5pAtoC_R | |
| Recombinant DNA reagent | pRmHa-mir-92b | This study | Genomic DNA fragment amplified by mir92b_genespecificF and mir92b_genespecificR inserted to pRmHa3 (DGRC: 1145) | |
| Recombinant DNA reagent | pRmHa-mir-92b A-to-U | This study | Site directed mutagenesis of pRmHa-mir-92b using dme_mir92b_5pAtoU_F and dme_mir92b_5pAtoU_R | |
| Recombinant DNA reagent | pRmHa-mir-92b A-to-G | This study | Site directed mutagenesis of pRmHa-mir-92b using dme_mir92b_5pAtoG_F and dme_mir92b_5pAtoG_R | |
| Recombinant DNA reagent | pRmHa-mir-92b A-to-C | This study | Site directed mutagenesis of pRmHa-mir-92b using dme_mir92b_5pAtoC_F and dme_mir92b_5pAtoC_R | |
| Recombinant DNA reagent | pRmHa-mir-263a | This study | Genomic DNA fragment amplified by mir263a_genespecifiF and mir263a_genespecifiR inserted to pRmHa3 (DGRC: 1145) | |
| Recombinant DNA reagent | pRmHa-mir-263a A-to-U | This study | Site directed mutagenesis of pRmHa-mir-263a using dme_mir263a_5pAtoU_F and dme_mir263a_5pAtoU_R | |
| Recombinant DNA reagent | pRmHa-mir-263a A-to-C | This study | Site directed mutagenesis of pRmHa-mir-263a using dme_mir263a_5pAtoC_F and dme_mir263a_5pAtoC_R | |
| Commercial assay or kit | Dual-Glo Luciferase Assay System | Promega | Promega:E2940 | |
| Commercial assay or kit | Effectene | QIAGEN | QIAGEN: 301427 | |
| Chemical compound, drug | 20-Hydroxyecdysone | Sigma | Sigma: H5142 | |
| Chemical compound, drug | Bathocuproinedisulfonic acid disodium salt | Sigma | Sigma: B1125 | |
| Software, algorithm | FASTX-toolkit | Hannon Lab | http://hannonlab.cshl.edu/fastx_toolkit | |
| Software, algorithm | Bowtie1.1.2 | PMID: 19261174 | ||
| Software, algorithm | STAR | PMID: 23104886 | ||
| Software, algorithm | Cufflinks suite tools | PMID: 20436464 | ||
| Software, algorithm | UCSC liftOver | UCSC Genome Browser | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
Additional files
-
Supplementary file 1
Library statistics (related to Figure 1).
Sheet 1: small RNA library mapping to dm3 genome. Sheet 2: small RNA library category mapping statistics. Sheet 3: small RNA library spike-in counts. Sheet 4: public total RNA-seq library used in this study. Sheet 5: public small RNA-seq library used in this study.
- https://doi.org/10.7554/eLife.38389.027
-
Supplementary file 2
miRNA expression level (related to Figure 1).
Sheet 1: miRNA Reads Counts (normalized by the number of genomic locations). Sheet 2: miRNA normalized reads (RPTS).
- https://doi.org/10.7554/eLife.38389.028
-
Supplementary file 3
Pri-miRNA transcription activity (related to Figure 1).
Sheet 1: Density of total RNA-seq reads in the upstream region of miRNA hairpin.
- https://doi.org/10.7554/eLife.38389.029
-
Supplementary file 4
Summary of multiple regression analysis (related to Figures 1 and 2).
- https://doi.org/10.7554/eLife.38389.030
-
Supplementary file 5
ANOVA analysis summary (related to Figure 3).
- https://doi.org/10.7554/eLife.38389.031
-
Supplementary file 6
DGRP polymorphic target analysis (related to Figure 7).
Sheet 1: Polymorphic target sites and their allele frequencies in the DGRP dataset. To draw the chart in Figure 7—figure supplement 1, polymorphic target sites were binned based on the allele frequency values (highlighted in red). Sheet 2: Counts of polymorphic target sites in each bin in Figure 7—figure supplement 1 charts. The fraction of polymorphic target sites (‘fraction’: highlighted in red) was used for the chart. Sheet 3: Derived target sites and their allele frequencies in the DGRP dataset. To draw the chart in Figure 7G, derived target sites were binned based on the derived allele frequency values (‘DAF’: highlighted in red). Sheet 4: Counts of derived target sites in each bin in Figure 7F charts. The fraction of derived target sites (‘fraction’: highlighted in red) was used for the chart.
- https://doi.org/10.7554/eLife.38389.032
-
Supplementary file 7
Oligos used in this study (related to Materials and methods).
- https://doi.org/10.7554/eLife.38389.033
-
Supplementary file 8
Genomic coordinates of additional isoforms of miRNA host transcripts (related to Materials and methods).
- https://doi.org/10.7554/eLife.38389.034
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38389.035





