Genetic basis for coordination of meiosis and sexual structure maturation in Cryptococcus neoformans
Figures
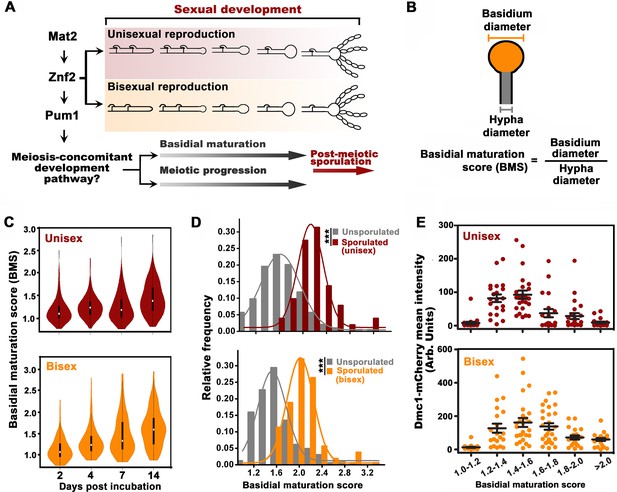
Basidial differentiation and meiotic progression are spatiotemporally coordinated in Cryptococcus neoformans.
(A) Diagram depicting hyphal development and meiosis-concomitant differentiation process in C. neoformans. (B) Schematic diagram outlining the basidial maturation score (BMS). (C) Violin plot analysis indicates that populations of basidia with high BMS gradually increased over time during unisexual and bisexual development. XL280α cells alone (unisex) or a mixture of XL280α and XL280a cells (α-a bisex) were dropped onto V8 medium and incubated at 25°C in the dark. Hyphae with or without basidia were photographed at 2, 4, 7 and 14 days after mating stimulation, and were randomly chosen for the BMS calculation (n > 110 for each time point). (D) Basidia were photographed at 7 days after mating stimulation. Unisex: n = 194 (unsporulated basidia); n = 51 (sporulated basidia). Bisex: n = 163 (unsporulated basidia); n = 53 (sporulated basidia). Bin width = 0.2. ***p<0.001, Kolmogorov-Smirnov test, two sided. A BMS of 1.0 was arbitrarily set as the threshold to define the basidial state. (E) Dynamic fluorescent intensity of Dmc1-mCherry during basidial maturation defined by the BMS method in cryptococcal unisex and bisex processes, respectively. n > 20 for each BMS range.
-
Figure 1—source data 1
Source file for Figure 1C,D and E.
- https://doi.org/10.7554/eLife.38683.006
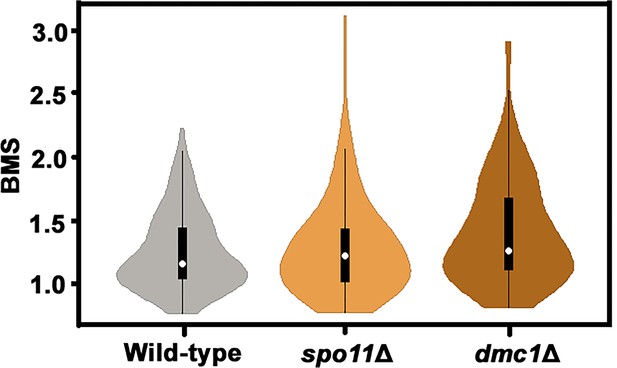
Violin plots showing the distribution frequency of the BMS in different strains.
α cells from XL280 and its derived mutants were incubated on V8 agar in the dark at 25°C to induce the unisexual mating response for 7 days. n = 118 for each strain.
-
Figure 1—figure supplement 1—source data 1
Source file for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.38683.005
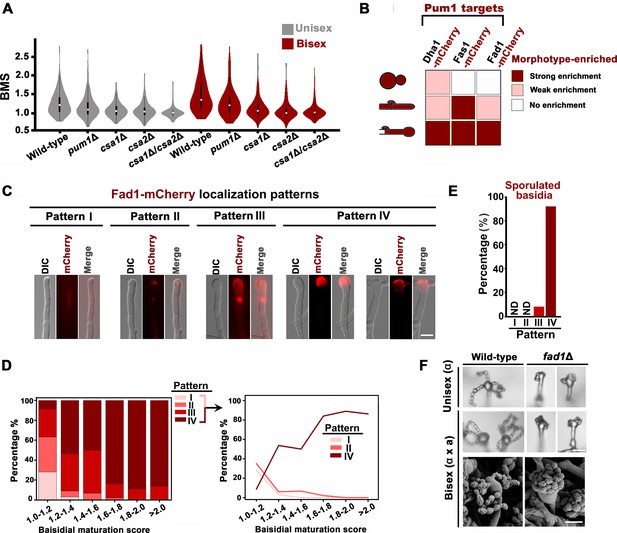
Pum1 orchestrates meiotic progression and basidial maturation.
(A) Violin plot analysis shows that disruption of PUM1 cascade members led to a decrease of high BMS basidial population during both unisexual and bisexual development (n = 150 for each strain). (B) Morphotype-specific enrichment of Dha1, Fas1, and Fad1. >50 cells in each morphotype were examined for mCherry-labelled proteins expression. (C) Dynamics of Fad1-mCherry expression during unisexual development. Fad1-mCherry shows a remarkably biased expression in the basidium structure and displays different localization patterns. Scale bar: 5 μm. (D) Cells were placed onto a V8 plate at 25°C in the dark for unisexual induction, and incubated for 7 days. For each BMS range, >20 basidia expressing Fad1-mCherry were examined. The right panel highlights the dynamic enrichment of patterns I, II and IV at various stages during basidial maturation. (E) Predominant Fad1 protein exhibited a subcellular localization identical to pattern IV in post-meiotic basidia (sporulated basidia) during unisexual reproduction. Thirty-seven sporulated basidia expressing Fad1-mCherry were measured. ND = Not Detected. (F) Sporulation phenotypes for wild-type XL280α, the fad1Δ deletion mutant (unisexual reproduction), a wild-type cross between XL280α and XL280a, and the fad1Δ bilateral mutant cross. Scale bar: 10 μm (upper and middle panels), 5 μm (bottom panels).
-
Figure 2—source data 1
Source file for Figure 2A,D and E.
- https://doi.org/10.7554/eLife.38683.009
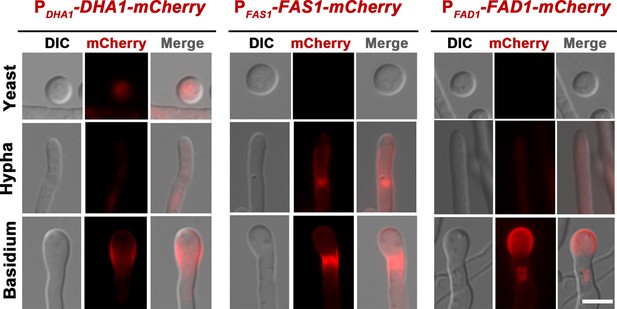
Morphotype-specific expression patterns of Dha1, Fas1 and Fad1, which are fused by mCherry at their C-terminus.
XL280α cells harboring genes encoding different mCherry-fused proteins were incubated on V8 agar for 7 days. Different morphotypes expressing mCherry-fused extracellular proteins were visualized. Representative images of n > 5 experiments. Scale bar: 5 μm.
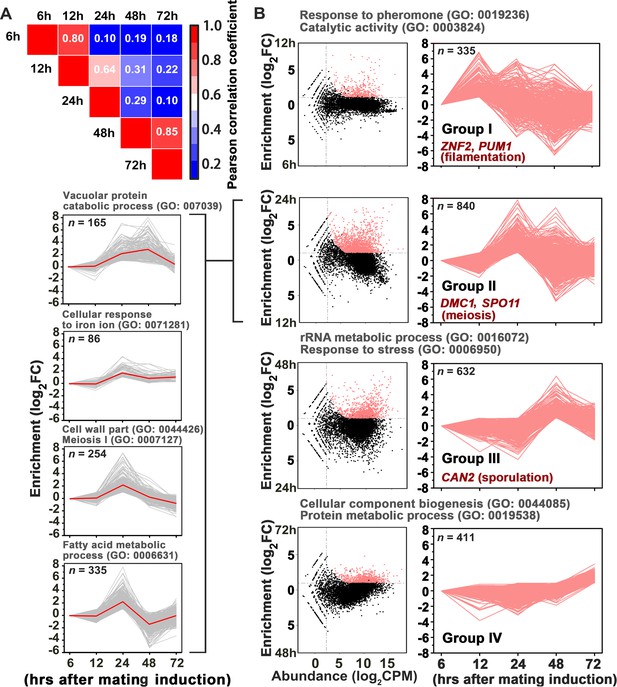
Gene network specifically orchestrating basidial maturation and meiosis during unisexual mating.
(A) Pairwise correlation of normalized TPM between RNA-seq samples obtained at five various time points after unisexual activation (6 hr, 12 hr, 24 hr, 48 hr and 72 hr). Pearson correlation coefficient was calculated using the R package ranges from no correlation (dark blue) to a perfect correlation (red). (B) Line plots show transcriptional induction profiles of genes in each pattern group. For each gene, the normalized expression levels at five time points throughout unisexual development are shown with a pink line (right). Pink dots indicate genes with significant induction during mating differentiation (CPM: count per million reads, FC: fold change). For each group, the representative genes, which play roles in various phases during sexual reproduction, are indicated (right). For each pattern from group II genes (left), the average expression levels at five time points across all genes with the pattern are shown with a red line. Tree cluster of group II genes was plotted using Cluster 3.0.
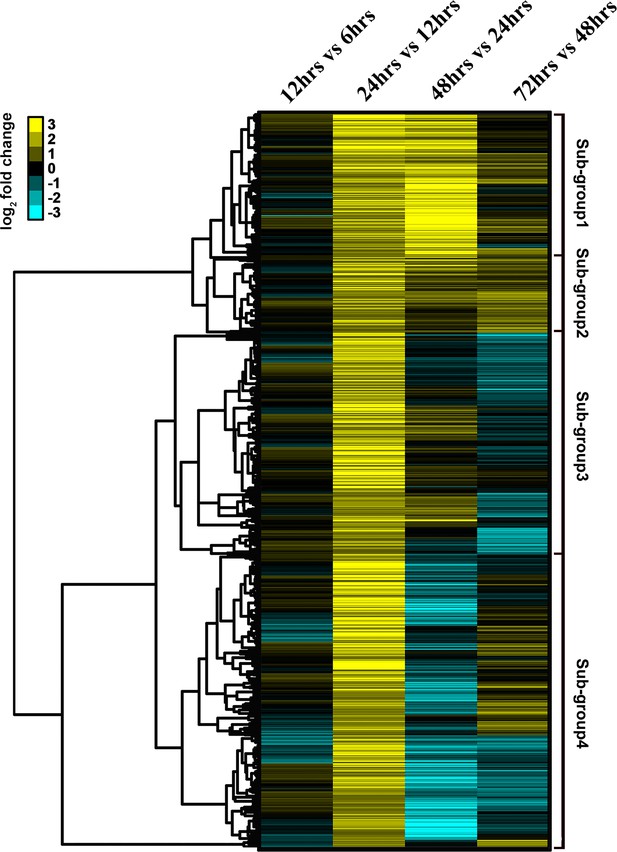
Group II genes are divided into four sub-groups using tree clustering (Cluster 3.0).
Color bar indicates the log2 fold change values.
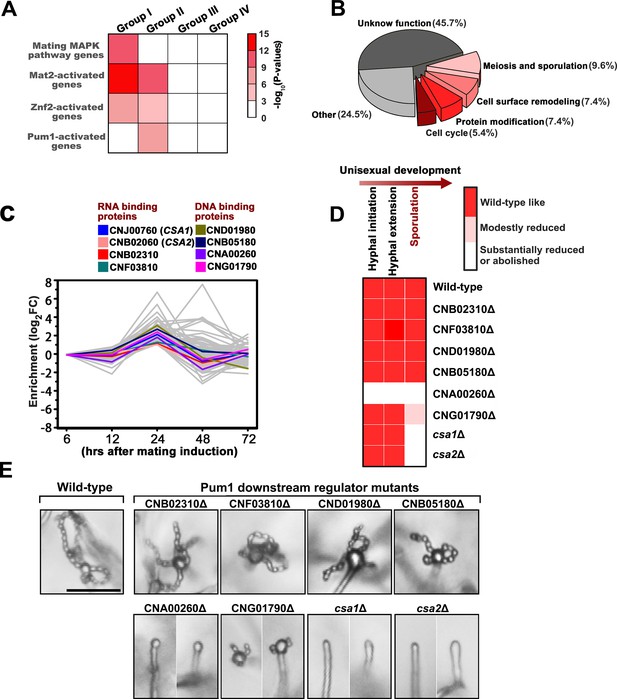
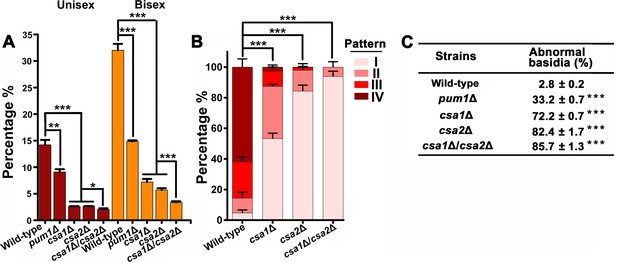
Csa1 and Csa2 as the key targets of Pum1 are required for post-meiotic sporulation.
(A) Enrichment of genes belonging to different signaling cascades in four gene groups. Among these mating cascades, only the set of genes activated by Pum1 was specifically enriched in group II. Genes used for the enrichment assessment include those encoding the published components (mating MAPK pathway) or the genes activated by the activators (Znf2, Mat2 and Pum1) dominating different sexual stages (Figure 4—source data 2). Genes activated by Mat2 and Znf2 (‘Mat2-activated genes’ and ‘Znf2-activated genes’) are derived from the previous transcriptome data (Lin et al., 2010). The gene set ‘Pum1-activated genes’ is generated based on the RNA-seq analysis of the PUM1 overexpression strain (Supplementary file 2). Only significantly enriched (p<0.001, Fisher’s exact test) families are colored. (B) Enriched GO terms of 94 group II genes induced by Pum1 using BiNGO. (C) Dynamic expression of the group II regulators with predicted RNA-binding or DNA-binding domains, whose mRNA levels were induced upon Pum1 overexpression, during unisexual reproduction. (D) Mating phenotypes for wild type XL280 and its isogenic mutant strains. Phenotype scores are represented in distinct colors based on quantitative or semi-quantitative analysis targeting the phenotypes related to sequential differentiation events during unisexual cycle. The results represent experiments from at least three independent mutants. (E) Sporulation phenotypes for XL280 (wild-type) and different Pum1 downstream regulator mutants during unisexual mating. Scale bar: 20 μm.
-
Figure 4—source data 1
Source file for Figure 4C.
The regulatory genes belonging to gene group II.
- https://doi.org/10.7554/eLife.38683.016
-
Figure 4—source data 2
Source file for Figure 4A.
Enrichment of the genes belonging to different signaling cascades in four gene groups.
- https://doi.org/10.7554/eLife.38683.017
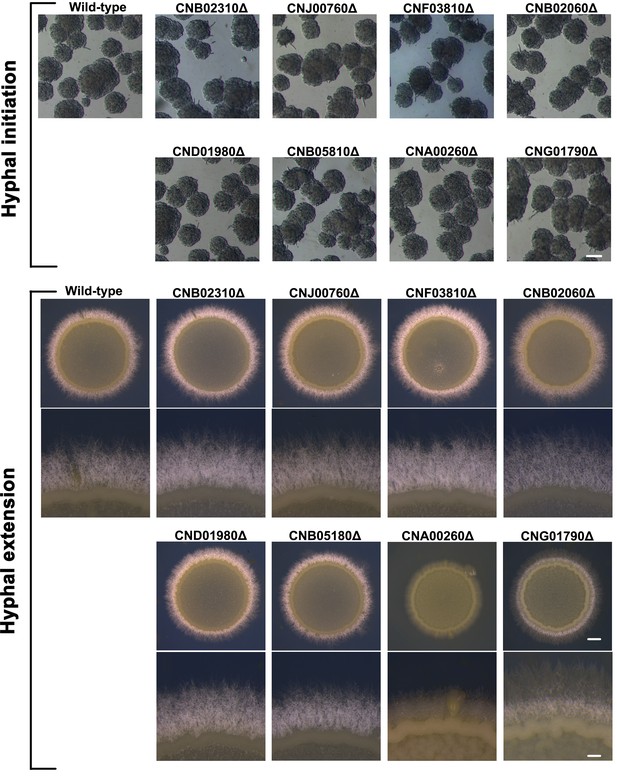
Mating phenotypes of the mutants lacking the regulators downstream of Pum1, including hyphal initiation and hyphal extension.
Scale bars: 100 μm (hyphal initiation); 1 mm (hyphal extension, upper panels) and 200 μm (hyphal extension, bottom panels).
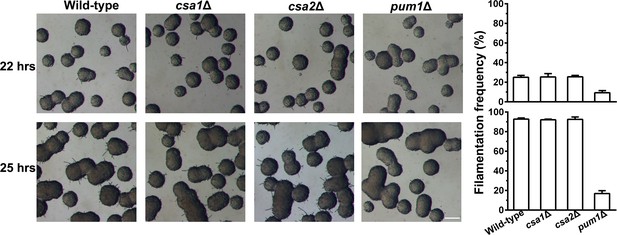
The absence of Pum1 but not its downstream targets Csa1 and Csa2 adversely affected self-filamentation during unisexual reproduction.
For quantitative analysis of self-filamentation frequency during unisex, the cells of each strain were plated onto V8 medium at a low cell density and allowed to grow into isolated mini-colonies after 22 hr or 25 hr of culture. Mini-colonies exhibited a great heterogeneity in filamentation (left), and filamentous incidence among mini-colonies reflects the strength of unisexual induction. Filamentation frequency (FF) is the percentage of filamentous mini-colonies. The dynamics in FF during unisexual development in different strains was calculated, respectively (right). n = 3 independent experiments, mean ± SD. Scale bar: 100 μm.
-
Figure 4—figure supplement 2—source data 1
Source file for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.38683.015
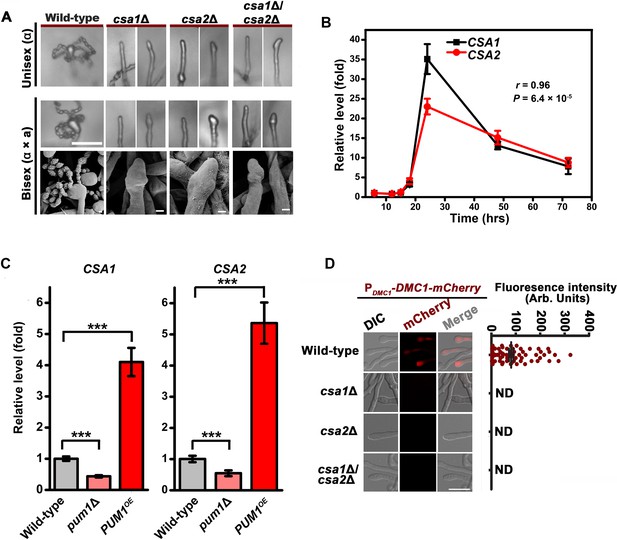
Csa1 and Csa2 govern the regulatory coordination of meiosis and basidial differentiation.
(A) Upper panels indicate sporulation phenotypes for wild-type XL280α, the csa1Δ mutant, the csa2Δ mutant and the csa1Δ/csa2Δ mutant during unisexual mating; middle and bottom panels illustrate sporulation phenotypes for a wild-type cross between XL280α and XL280a, the csa1Δ bilateral mutant cross, the csa2Δ bilateral mutant cross, and the csa1Δ/csa2Δ bilateral mutant cross. Scale bar: 20 μm (upper and middle panels). Scale bar: 1 μm (bottom panels). (B) RT-PCR analysis showed the dynamic expression of CSA1 and CSA2 at seven different time points (6 hr, 12 hr, 15 hr, 18 hr, 24 hr, 48 hr and 72 hr) during unisexual mating. Bars show the mean ±SD of six individual experiments. (C) RT-PCR analysis indicated that the mRNA levels of both CSA1 and CSA2 were positively affected by PUM1 during unisexual reproduction at 24 hr post inoculation on mating inducing V8 medium. Bars show the mean ± SD of six individual experiments. ***p<0.001, two-tailed Student’s t-test. (D) The images of the fluorescence-labeled strains were taken at 7 days after incubation on V8 medium (left). > 50 basidia for each strain were examined for the expression of Dmc1-mCherry (right). Scale bar: 10 µm. ND = Not Detected.
-
Figure 5—source data 1
Source file for Figure 5B,C and D.
- https://doi.org/10.7554/eLife.38683.021
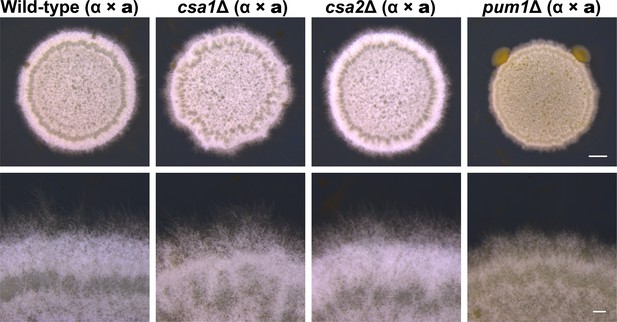
Deletion of PUM1 but not CSA1 and CSA2 attenuated bisexual filamentation in bilateral mating assays.
Bisexual filamentation for a wild-type cross between XL280α and XL280a, pum1Δ, csa1Δ and csa2Δ bilateral mutant crosses. All mating patches were spotted on V8 medium and incubated in the dark at 25°C for 7 days. Scale bars: 1 mm (upper) and 200 μm (bottom).
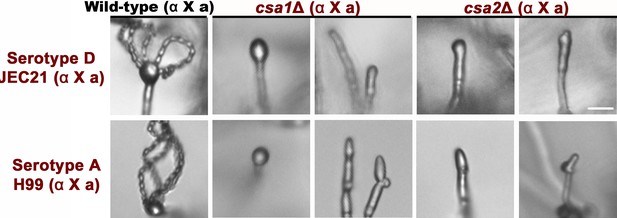
Deletion of CSA1 and CSA2 blocked bisexual sporulation in both laboratory serotype D strain (JEC21) and clinical serotype A isolate (H99).
Bisexual wild type crosses (JEC21 α ×JEC20 a) and bilateral mutant crosses (csa1Δα ×csa1Δa, csa2Δα ×csa2Δa) were conducted on V8 medium in the dark at 25°C to stimulate matings. Sporulation phenotypes were photographed at 7 days (serotype D) or 1 month (serotype A) after mating stimulation. Scale bar = 10 μm.
Csa1 and Csa2 can function in parallel in basidial maturation and morphogenesis.
(A) Compared with either of the single deletion, the csa1Δ/csa2Δ mutant displayed a lower number of mature basidia (BMS >1.6) during both unisexual and bisexual reproduction. In both reproduction modes, >150 basidia were examined for each test. Bars show the mean ±SD of three independent experiments. ***p<0.001, *p<0.05, two-tailed Student’s-t test. (B) Cells were placed onto V8 plate and incubated in the dark at 25°C for the unisexual simulation. >150 basidia for each strain expressing Fad1-mCherry were visualized. Bars show the mean ±SD of three independent experiments. ***p<0.001, two-tailed Student’s-t test. (C) α cells from XL280 and its derived mutants were incubated on V8 agar in the dark at 25°C to induce the unisexual mating response. Basidia were photographed at 7 days after incubation. >100 basidia for each strain were tested. Data are presented as the mean ± SD from three independent experiments. ***p<0.001 indicates the significant difference compared to the wild-type strain, two-tailed Student’s-t test.
-
Figure 6—source data 1
Source file for Figure 6A and B.
- https://doi.org/10.7554/eLife.38683.029
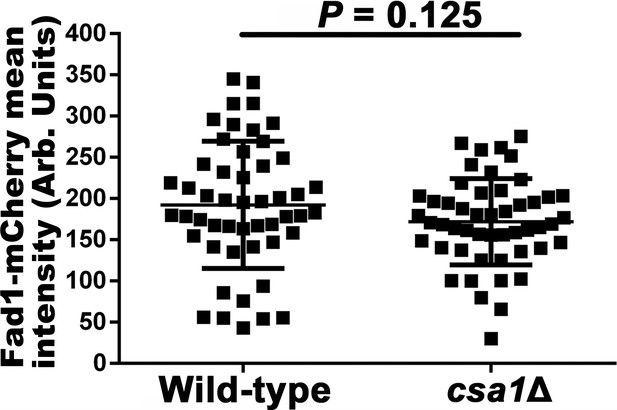
Fad1-mCherry expression was not significantly affected in the absence of Csa1.
50 basidia expressing Fad1-mCherry were examined for each strain at 7 days post mating induction during unisex. Statistical significance was defined using two-tailed Student’s t test.
-
Figure 6—figure supplement 1—source data 1
Source file for Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.38683.024
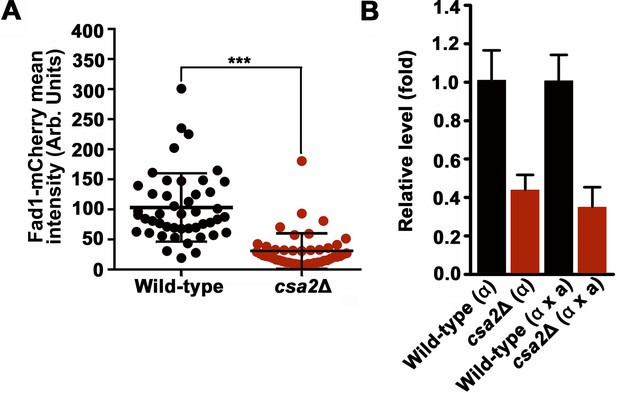
Fad1 is a downstream target of Csa2.
(A) Deletion of CSA2 led to a significant reduction in the protein level of Fad1-mCherry after 7 days incubation on mating-inducing V8 medium. Asterisks indicate statistical significance calculated using a two-tailed Student’s-t test. >40 basidia were tested for each strain. (B) RT-PCR analysis showed that the FAD1 mRNA level was down-regulated in the csa2Δ mutant at 48 hr post mating induction during both unisexual and bisexual reproduction. Bars show the mean ±SD of four individual experiments.
-
Figure 6—figure supplement 2—source data 1
Source file for Figure 6—figure supplement 2A and B.
- https://doi.org/10.7554/eLife.38683.026
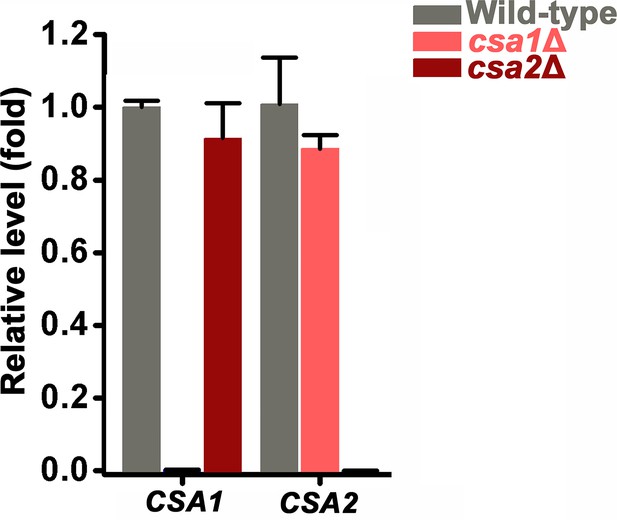
RT-PCR analysis indicated that CSA1 and CSA2 do not appear to affect the expression of each other at 48 hr post inoculation on mating inducing V8 medium.
Bars show the mean ±SD of three individual experiments.
-
Figure 6—figure supplement 3—source data 1
Source file for Figure 6—figure supplement 3.
- https://doi.org/10.7554/eLife.38683.028
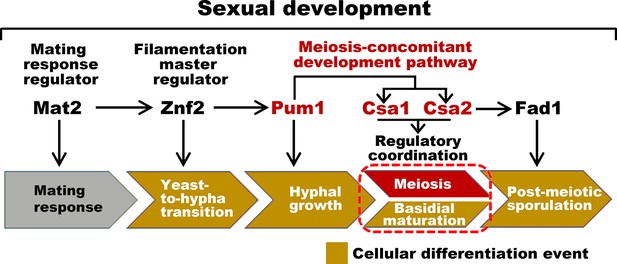
Sexual control in C.neoformans.
Model describing the genes responsible for sequential events during sexual reproduction. Csa1 and Csa2 governs the regulatory coordination of basidial maturation and meiosis, which is required for sporulation.
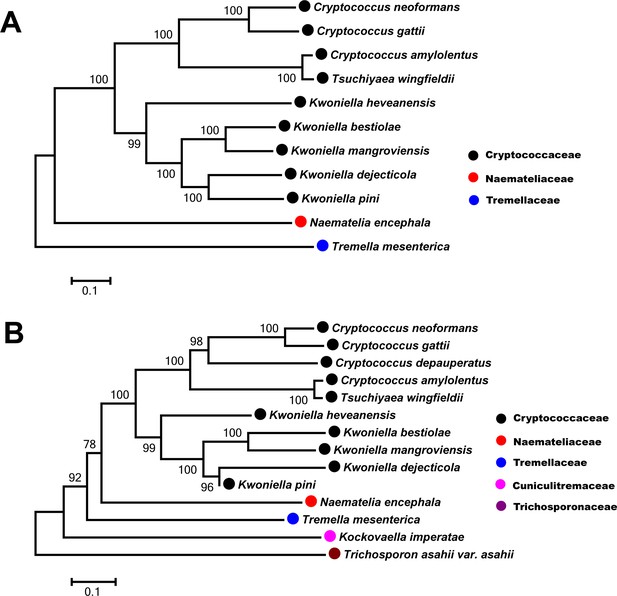
Phylogenetic tree of Csa1 orthologs.
(A) and Csa2 orthologs (B) based on amino acid sequence aligned using the neighbor-joining method with the MEGA v7.0.18 program. Homologues of Csa1 and Csa2 with greater than 30% identity and greater than 50% coverage were selected to construct the phylogenetic tree.
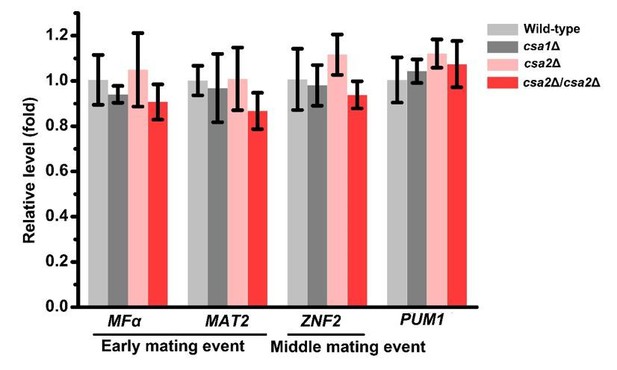
RT-PCR analysis showed that deleting either or both of CSA1 and CSA2 cannot significantly change the mRNA levels of MFα, MAT2, ZNF2 and PUM1 at 24 hrs post unisexual mating stimulation.
Bars show the mean ± SD of three replicates.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (C. neoformans species complex) | XL280, MATα, wild-type | PMID: 17112316 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, wild-type | PMID: 23670559 | ||
| Genetic reagent (C. neoformans species complex) | JEC21, MATα, wild-type | PMID: 10512666 | ||
| Genetic reagent (C. neoformans species complex) | JEC20, MATa, wild-type | PMID: 10512666 | ||
| Genetic reagent (C. neoformans species complex) | KN99, MATα, wild-type | PMID: 12933823 | ||
| Genetic reagent (C. neoformans species complex) | KN99, MATa, wild-type | PMID: 12933823 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA2::NEOr | This study | See Materials and methods, ‘Gene disruption and overexpression’ | |
| Genetic reagent (C. neoformans species complex) | XL280, MATa, CSA2::NEOr | This study | ||
| Genetic reagent(C. neoformans species complex) | H99, MATα, CSA2::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | H99, MATa, CSA2::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | JEC21, MATα, CSA2::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | JEC20, MATa, CSA2::NEOr | This study | ||
| Genetic reagentC. neoformans species complex) | XL280, MATα, FAD1::NEOr | PMID: 24901238 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, FAD1::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PUM1::NEOr | PMID: 24901238 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, PUM1::NEOr | PMID: 24901238 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, CSA1::NATr | This study | ||
| Genetic reagent (C. neoformans species complex) | H99, MATα, CSA1::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | H99, MATa, CSA1::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | JEC21, MATα, CSA1::NEOr | This study | ||
| gGenetic reagent (C. neoformans species complex) | JEC20, MATa, CSA1::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, DMC1::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, SPO11::NEOr | PMID: 23966871 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA2::NEOr, CSA1::NATr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, CSA2::NEOr, CSA1::NATr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PRPBL2B-PUM1-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PDMC1-DMC1-mCherry-3'UTR-HYG | PMID: 24901238 | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATa, PDMC1-DMC1- mCherry-3'UTR-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA2::NEOr, PDMC1- DMC1-mCherry-3'UTR-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr, PDMC1- DMC1-mCherry-3'UTR-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PFAD1-FAD1-mCherry- HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA2::NEOr, PFAD1-FAD1-mCherry-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PFAS1-FAS1-mCherry-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, PDHA1-DHA1- mCherry-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr, PFAD1- FAD1-mCherry-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr, PFAD1- FAD1-mCherry-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr, CSA2::NEOr, PDMC1-DMC1-mCherry-3'UTR-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CSA1::NATr, CSA2::NEOr, PFAD1-FAD1-mCherry-3'UTR-HYG | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNB02310::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNF03810::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNF01980::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNB05180::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNF00260::NEOr | This study | ||
| Genetic reagent (C. neoformans species complex) | XL280, MATα, CNG01790::NEOr | This study | ||
| Software, algorithm | RStudio Version 1.1.456 | RStudio | RRID:SCR_000432 | |
| Software, algorithm | FastQC v0.11.5 | RRID:SCR_014583 | ||
| Software, algorithm | DEseq2 v1.16.1 | RRID:SCR_016533 | ||
| Software, algorithm | Hisat2 v2.1.0 | RRID:SCR_015530 | ||
| Software, algorithm | BiNGO v3.0.3 | RRID:SCR_005736 | ||
| Software, algorithm | Graphpad Prism 6 | Graphpad | RRID:SCR_002798 | |
| Sequence- based reagent | Wanglab959 (knockout primer pairs) | This study | TTGTCACCAACCTATCCGCTAC | |
| Sequence- based reagent | Wanglab960 (knockout primer pairs) | This study | CAGTTTGCTCTTATTCCCACTCC | |
| Sequence- based reagent | Wanglab961 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGAAGCACTTGGTGAATGAGACATT | |
| Sequence- based reagent | Wanglab962 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGCACCGCCCTTACGATTATACATCT | |
| Sequence- based reagent | Wanglab963 (knockout primer pairs) | This study | GTTGTGAGTGTCATGAGTGTCATTG | |
| Sequence- based reagent | Wanglab964 (knockout primer pairs) | This study | CCTCTTCTGCCAATAACCCTTTT | |
| Sequence- based reagent | Wanglab2195 (knockout primer pairs) | This study | CATCCCCAGAACACGCTGAT | |
| Sequence- based reagent | Wanglab2196 (knockout primer pairs) | This study | TCCGGCCATTAAGATCCGTG | |
| Sequence- based reagent | Wanglab2197 (knockout primer pairs) | This study | AAAACGGCAACAGTCAAGGC | |
| Sequence- based reagent | Wanglab2198 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGTTGTTAAAGGCAGTTGAGCGA | |
| Sequence- based reagent | Wanglab2199 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGAAGGCATCACTTCGTTTGGC | |
| Sequence- based reagent | Wanglab2200 (knockout primer pairs) | This study | AACCATAGGATGTGCCACGC | |
| Sequence- based reagent | Wanglab953 (knockout primer pairs) | This study | CCGTAGGCTTATCCCAGTCAGA | |
| Sequence- based reagent | Wanglab954 (knockout primer pairs) | This study | GTGGAAGGCAAGAGTTGGTGTT | |
| Sequence- based reagent | Wanglab955 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACACATTTCCAGAAGAGGCAAGAAGA | |
| Sequence- based reagent | Wanglab956 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGGGTAGAAGAACGTCAAACAACTAA | |
| Sequence- based reagent | Wanglab957 (knockout primer pairs) | This study | CCTTGGCAACAGTAGGCTTCTG | |
| Sequence- based reagent | Wanglab958 (knockout primer pairs) | This study | GGAAGGGAGTGGTGAGGTTGAA | |
| Sequence- based reagent | Wanglab2461 (knockout primer pairs) | This study | GGGCCTGAAAAGTATGAAGTCC | |
| Sequence- based reagent | Wanglab2462 (knockout primer pairs) | This study | TAGCCTTTCCACCACAGCAGC | |
| Sequence- based reagent | Wanglab2463 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGTAGCGGTTTCGACGGACATAT | |
| Sequence- based reagent | Wanglab2464 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGGAAGAGGAGGAGACCAAGGAG | |
| Sequence- based reagent | Wanglab2465 (knockout primer pairs) | This study | ATCCTTTGTCCAACCCGTGAG | |
| Sequence- based reagent | Wanglab2466 (knockout primer pairs) | This study | GCCCATGTCGCATTACGTAAAG | |
| Sequence- based reagent | Wanglab2423 (knockout primer pairs) | This study | AGCCATTCGGCTCTTATCGC | |
| Sequence- based reagent | Wanglab2424 (knockout primer pairs) | This study | AGCGACTGCGACCATTATGT | |
| Sequence- based reagent | Wanglab2425 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACATGGAGGCGTTGGAGAATCC | |
| Sequence- based reagent | Wanglab2426 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGCAAGACGTGCATACCCTCTA | |
| Sequence- based reagent | Wanglab2427 (knockout primer pairs) | This study | GCTTCAGTATGCCAACCCCT | |
| Sequence- based reagent | Wanglab2428 (knockout primer pairs) | This study | CGAGAGAAGGGAAAGCGAGG | |
| Sequence- based reagent | Wanglab2201 (knockout primer pairs) | This study | GGAGAGATCAGAGGCAGCAC | |
| Sequence- based reagent | Wanglab2202 (knockout primer pairs) | This study | CGTCGTGGAAAAGGTGAGGA | |
| Sequence- based reagent | Wanglab2203 (knockout primer pairs) | This study | TCCGGATTTCTCAAGTGGGC | |
| Sequence- based reagent | Wanglab2204 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGCT CTAGCATTTGCGGGGAT | |
| Sequence- based reagent | Wanglab2205 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGTGACTCCCCCTCCAGAAAGC | |
| Sequence- based reagent | Wanglab2206 (knockout primer pairs) | This study | AACCAAAATGGCTCCGGACA | |
| Sequence- based reagent | Wanglab2682 (knockout primer pairs) | This study | TTGCAACCATCCGAGGTCAA | |
| Sequence- based reagent | Wanglab2683 (knockout primer pairs) | This study | GAAATCCGACACCTCCCTGG | |
| Sequence- based reagent | Wanglab2684 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGG GATGTTTGTCCCTTTCGC | |
| Sequence- based reagent | Wanglab2685 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGACCAGTAAGAAGCGGTGACA | |
| Sequence- based reagent | Wanglab2686 (knockout primer pairs) | This study | AGCGCTCGACTAGCTTTCTC | |
| Sequence- based reagent | Wanglab2687 (knockout primer pairs) | This study | GGATCCAAGACCTCCGATGG | |
| Sequence- based reagent | Wanglab3060 (knockout primer pairs) | This study | AGCGATAAGCCAGCAAGAGTT | |
| Sequence- based reagent | Wanglab3061 (knockout primer pairs) | This study | CCTCGAACCCGATACTGACG | |
| Sequence- based reagent | Wanglab3062 (knockout primer pairs) | This study | AGCTTAGAATAGCGACCGCC | |
| Sequence- based reagent | Wanglab3063 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACTGTGA GAGTCGGCTGATAGGA | |
| Sequence- based reagent | Wanglab3064 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGTGGAACCTAATTGCACCGC | |
| Sequence- based reagent | Wanglab3065 (knockout primer pairs) | This study | ATGGCGAGTTGCTTTCATGC | |
| Sequence- based reagent | Wanglab3066 (knockout primer pairs) | This study | TAATGTCGCTGAAGGGCCTG | |
| Sequence- based reagent | Wanglab3067 (knockout primer pairs) | This study | CCAAGGGTCAGCTATCCAGC | |
| Sequence- based reagent | Wanglab3068 (knockout primer pairs) | This study | CCGTAACCGGTGAGACATCA | |
| Sequence- based reagent | Wanglab3069 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGAGACGAATGAGCTGTGGCA | |
| Sequence- based reagent | Wanglab3070 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGTCAA GTCATGCCTGTGATCCT | |
| Sequence- based reagent | Wanglab3071 (knockout primer pairs) | This study | AGATCCTGGAGGGAACGGAT | |
| Sequence- based reagent | Wanglab3072 (knockout primer pairs) | This study | TTAGCTCGCCCTCGCTTATT | |
| Sequence- based reagent | Wanglab3073 (knockout primer pairs) | This study | AGCCAACCCATTTACCGACT | |
| Sequence- based reagent | Wanglab3074 (knockout primer pairs) | This study | CGTTGGACAATGGAGTGAGGA | |
| Sequence- based reagent | Wanglab3075 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGGGGA TGAAGGGAGCTAAAGG | |
| Sequence- based reagent | Wanglab3076 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGAAG CCTTTGCATTTGACCCT | |
| Sequence- based reagent | Wanglab3077 (knockout primer pairs) | This study | GGACAGAGGCCGTCAACATA | |
| Sequence- based reagent | Wanglab3646 (knockout primer pairs) | This study | CTAACGACAACAAGAAACCACGAC | |
| Sequence- based reagent | Wanglab3647 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACAGGCGGA GGAAGGTAGGAGAA | |
| Sequence- based reagent | Wanglab3648 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGTAGGTAA TGTTGACGGTGGTGA | |
| Sequence- based reagent | Wanglab3649 (knockout primer pairs) | This study | GTCTTAGTGGTCTGAGCCGAATAC | |
| Sequence- based reagent | Wanglab3650 (knockout primer pairs) | This study | AGGACGCTATTCGCTCTATCGG | |
| Sequence- based reagent | Wanglab3651 (knockout primer pairs) | This study | GATCCTTCACCCTGACTCTGTTCA | |
| Sequence- based reagent | Wanglab3261 (knockout primer pairs) | This study | ACTCATGCCTACCCATTGCC | |
| Sequence- based reagent | Wanglab3262 (knockout primer pairs) | This study | GCGACTCACTGAGCTTGACA | |
| Sequence- based reagent | Wanglab3263 (knockout primer pairs) | This study | CGGGCTTTACACCTACTCGG | |
| Sequence- based reagent | Wanglab3264 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACTCTGC TTGTACGTCAGCGAT | |
| Sequence- based reagent | Wanglab3265 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGAGTGA AGAGACTTGACGCTCG | |
| Sequence- based reagent | Wanglab3266 (knockout primer pairs) | This study | ACTAGCCCGAAGTGATGGGA | |
| Sequence- based reagent | Wanglab3267 (knockout primer pairs) | This study | GGCGCGTTGTAAAGCAGTAG | |
| Sequence- based reagent | Wanglab3268 (knockout primer pairs) | This study | TCTCCCCTCGGAAACAGCTA | |
| Sequence- based reagent | Wanglab3269 (knockout primer pairs) | This study | AGCACCTTTGCGATGTCTGA | |
| Sequence- based reagent | Wanglab3270 (knockout primer pairs) | This study | CTGGCCGTCGTTTTACGTTC CTGGACCCTTGATCCC | |
| Sequence- based reagent | Wanglab3271 (knockout primer pairs) | This study | GTCATAGCTGTTTCCTGGC AGTAACGGTCCTGTTCCA | |
| Sequence- based reagent | Wanglab3272 (knockout primer pairs) | This study | GTTCGATCAGAAACACGGCG | |
| Sequence- based reagent | Wanglab857 (qRT-PCR primer) | This study | CGTCACCACTGAAGTCAAGT | |
| Sequence- based reagent | Wanglab858 (qRT-PCR primer) | This study | AGAAGCAGCCTCCATAGG | |
| Sequence- based reagent | Wanglab3401 (qRT-PCR primer) | This study | AGACTCGACCACAGGCAG | |
| Sequence- based reagent | Wanglab3402 (qRT-PCR primer) | This study | AAAGGACAGGGTCAGGGTT | |
| Sequence- based reagent | Wanglab2583 (qRT-PCR primer) | This study | TTCTGCCGTAATGGGTGTCA | |
| Sequence- based reagent | Wanglab2584 (qRT-PCR primer) | This study | TCGTAAGGGCGGTGTTGTG | |
| Sequence- based reagent | Wanglab2585 (qRT-PCR primer) | This study | GTGAGATTATTGCCCGTGATGA | |
| Sequence- based reagent | Wanglab2586 (qRT-PCR primer) | This study | TTGGAGACGCCAGGGATGT | |
| Sequence- based reagent | Wanglab855 | This study | CTCTGGTTGGCACGGTG | |
| Sequence- based reagent | Wanglab856 | This study | CGTCGGTCAATCTTCTCG | |
| Sequence- based reagent | Wanglab2689 (overexpression primer) | This study | TTTGCATTGCGGCCGCAGGG GTGAATCGATATTCGACGC | |
| Sequence- based reagent | Wanglab2690 (overexpression primer) | This study | GGATAATTGCGATCGCCAGCTG GAGAGTGACAGACTTGG |
Additional files
-
Supplementary file 1
Expression profiles of various developmental stages during unisexual reproduction.
- https://doi.org/10.7554/eLife.38683.032
-
Supplementary file 2
Expression profiles of PUM1OE during unisexual reproduction.
- https://doi.org/10.7554/eLife.38683.033
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38683.034





