Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5
Figures
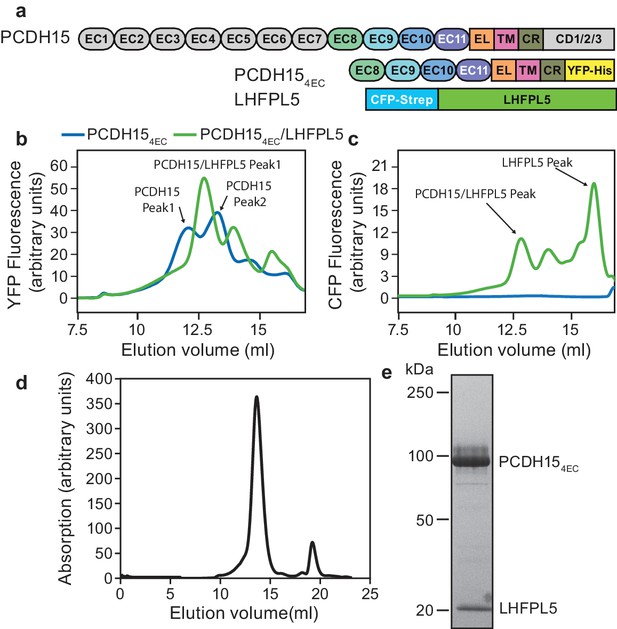
PCDH15 and LHFPL5 form a stable and monodisperse complex.
(a) Schematic of constructs used for interaction screening of PCDH15 and LHFPL5 by mcFSEC. (b,c) mcFSEC analysis of lysates from cells expressing either PCDH154EC alone or together with LHFPL5. Elution of PCDH154EC is monitored by YFP fluorescence (b) and elution of LHFPL5 is measured by CFP fluorescence (c). (d) Preparative SEC trace of the PCDH154EC/LHFPL5 complex as monitored by absorbance at 280 nm. (e) SDS-PAGE analysis of the PCDH154EC/LHFPL5 complex following SEC purification stained using coomassie.
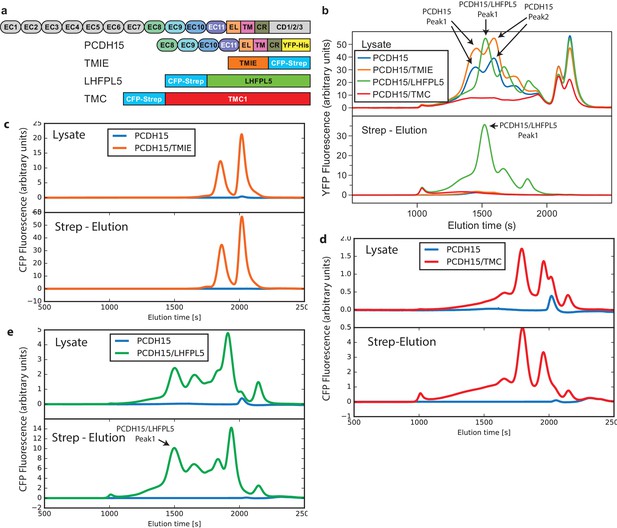
FSEC analysis of lysates and affinity purification eluates from PCDH15 coexpression screen.
(a) Schematic of used constructs. (b) Lysates and eluates analyzed by mcFSEC using the YFP fluorescence. (c,d,e), Lysates and eluates analyzed by mcFSEC using the CFP fluorescence in lysates where PCDH15 was coexpressed with TMIE (c), TMC (d), and LHFPL5 (e).
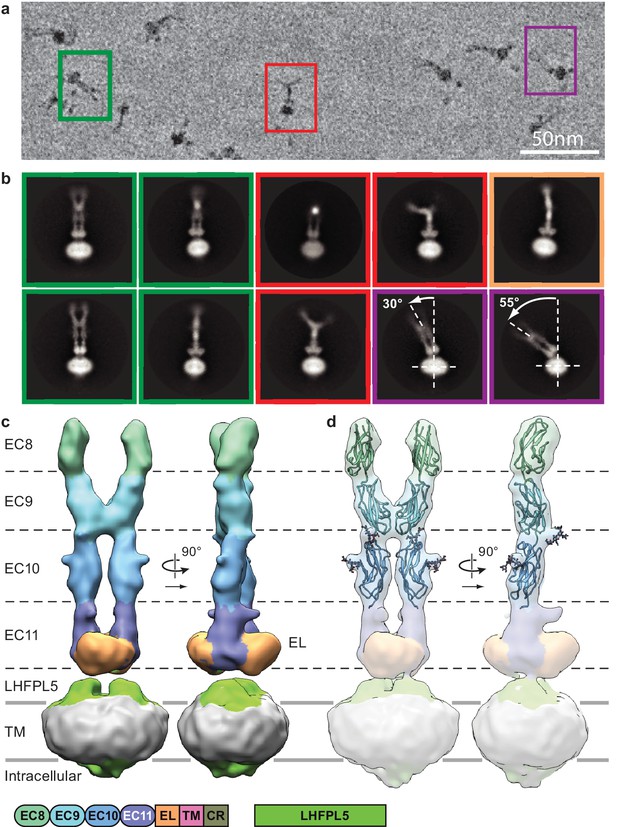
Cryo-EM analysis of PCDH154EC/LHFPL5 complex.
(a) Representative subsection of a micrograph containing particles of the straight conformation (green), split conformation (red), and tilted conformation (purple). (b) Representative 2D classes. Classes are color coded according to panel (a) with the addition of a bend conformation (gold). (c) 3D reconstruction of the straight conformation. (d) Fit of individual domains from PDB structure 4XHZ into the density. Schematic of the corresponding constructs is listed at the bottom.
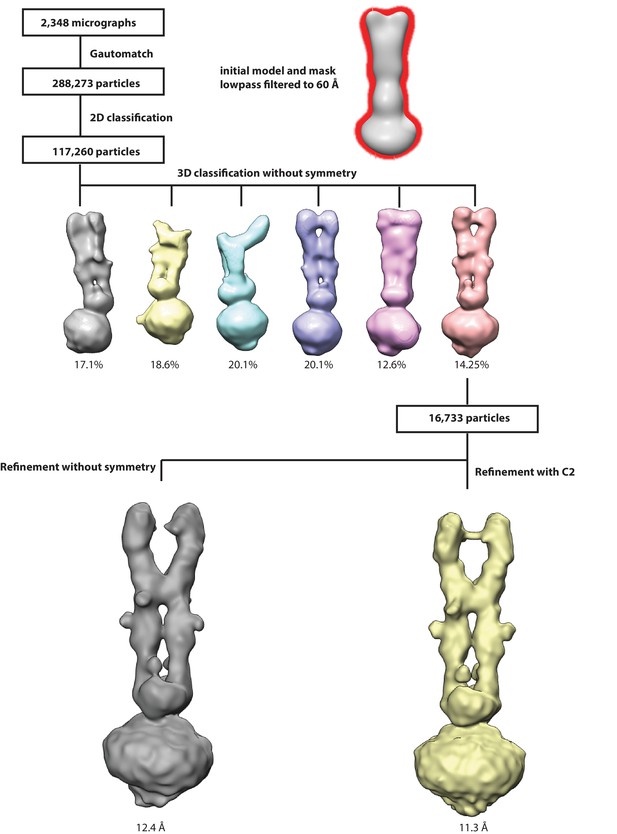
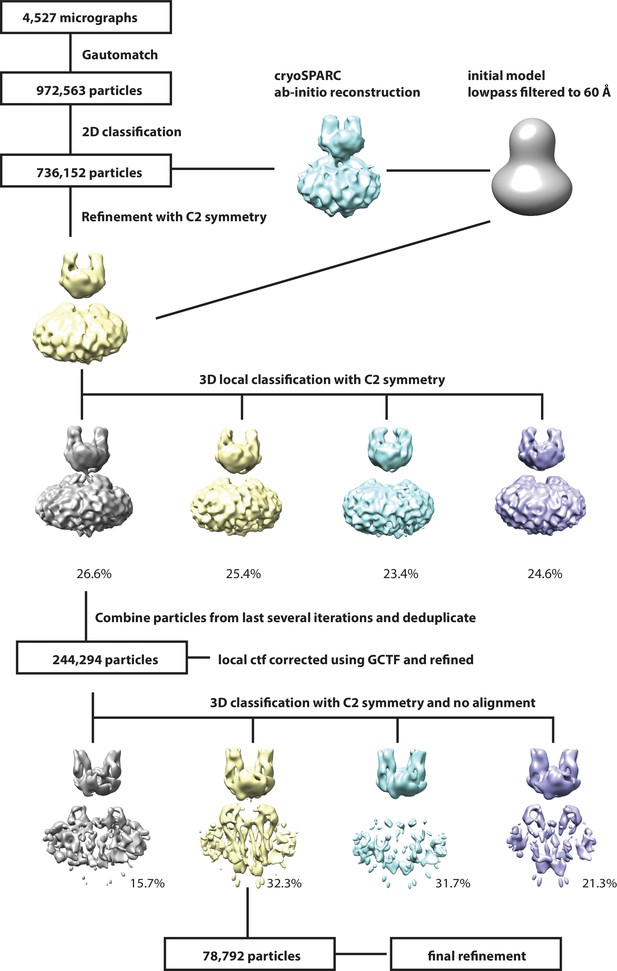
Workflow for 3D reconstruction of PCDH154EC/LHFPL5 complex.
https://doi.org/10.7554/eLife.38770.006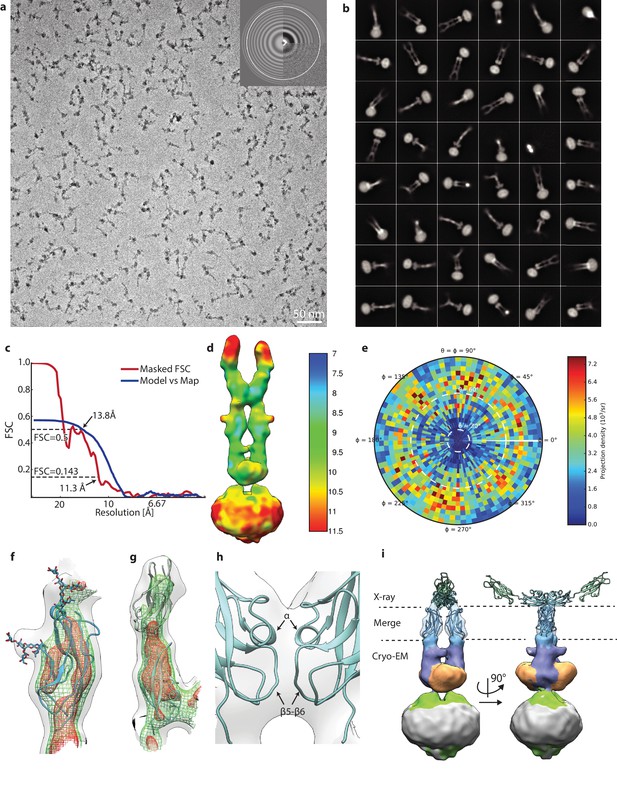
Details of PCDH154EC/LHFPL5 cryo-EM.
(a) Representative micrograph. (b) 48 most populated 2D class averages. (c) FSC analysis of final reconstruction. (d) Local resolution estimate. (e) Angular distribution. (f,g) Details of fitting EC10 (f) and EC8-9 (g). Map is shown at various thresholds with transparent grey surface, green mesh, and red mesh. (h) Details of EC9 dimer interface. (i) X-ray structure of 4XHZ fit into map using only EC10. Cryo-EM density for EC8-9 is hidden for clarity.
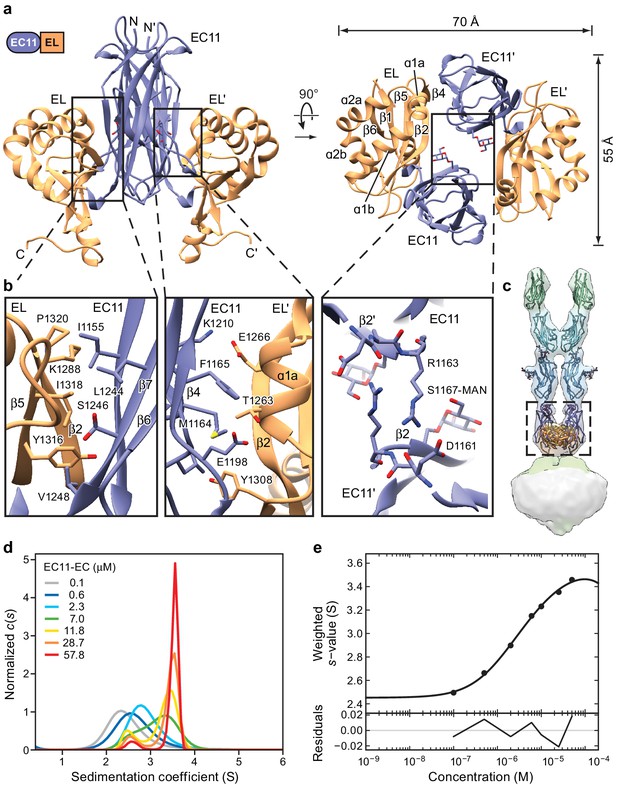
Crystal structure of the PCDH15 EC11-EL dimer.
(a) Ribbon representation of the crystal structure. Mannose modification at Ser1167 is shown in stick representation. (b) Detailed view of interactions mediating dimer formation. (c) Fit of the PCDH15 EC11-EL crystal structure into the density of the PCDH154EC/LHFPL5 complex. (d, e) AUC analysis of the PCDH15 EC11-EL. (d) Concentration dependence of the c(s) distributions with loading concentrations between 0.1–57.8 μM. (e) Isotherm of signal-weighted average sedimentation coefficient, sw, from the c(s) distributions shown in (d). The best-fit value of KD is 5.7 μM (95% confidence intervals: 3.3–9.9 μM). Best-fit values for monomer and dimer sedimentation coefficients are 2.46 S (95% confidence intervals: 2.37–2.54 S) and 3.74 S (95% confidence intervals: 3.63–3.89 S), respectively.
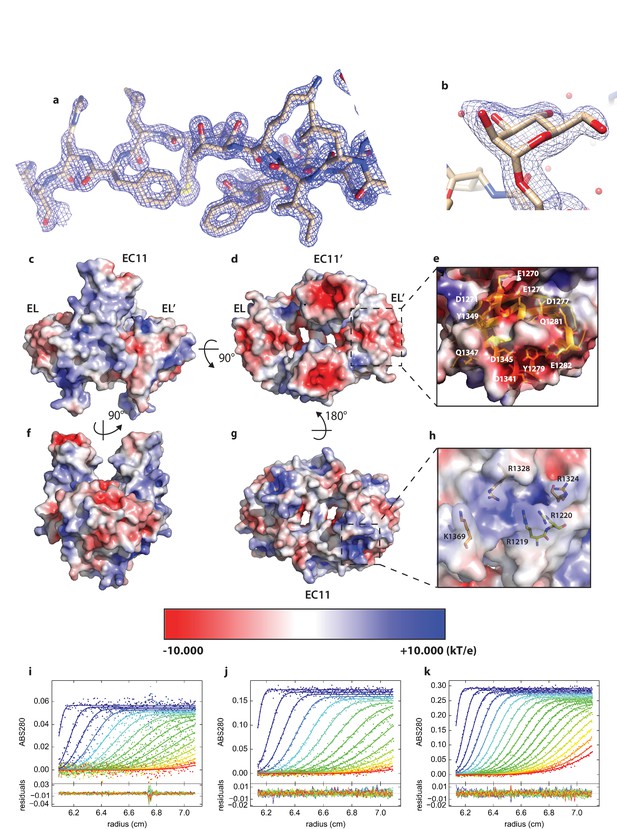
Details of PCDH15_EC11-EL crystal structure.
(a) Representative area of structure with 2mFo-DFc map at 2.1 I/σ. b, Glycosylation at S1167 modeled as α-Mannose. (c,d,e,f,g,h) Electrostatics analysis of PCDH15 EC11-EL. (c,d,f,g) Electrostatic potential of EC11-EL’s surface in various views. (e), Residues that contribute to the negatively charged region indicated in (d). (h) Residues that contribute to the positively charged region indicated in (g). (i,j,k) Global boundary modeling of AUC data with Lamm equation solutions using the kinetic homo-dimerization model for protocadherin 15 at 2.3 µM (i), 7.0 µM (j) and 11.8 µM (k). For clarity, only every 3rd data point of every 2nd scan is shown (filled circles, using color temperature blue to red to indicate progression of scan time), with the solid lines representing the corresponding best-fit model, and residuals plotted in the bottom in each panel. The best-fit value of koff is 3.5×10−4 sec−1 and KD is 6.64 µM from the global analysis.
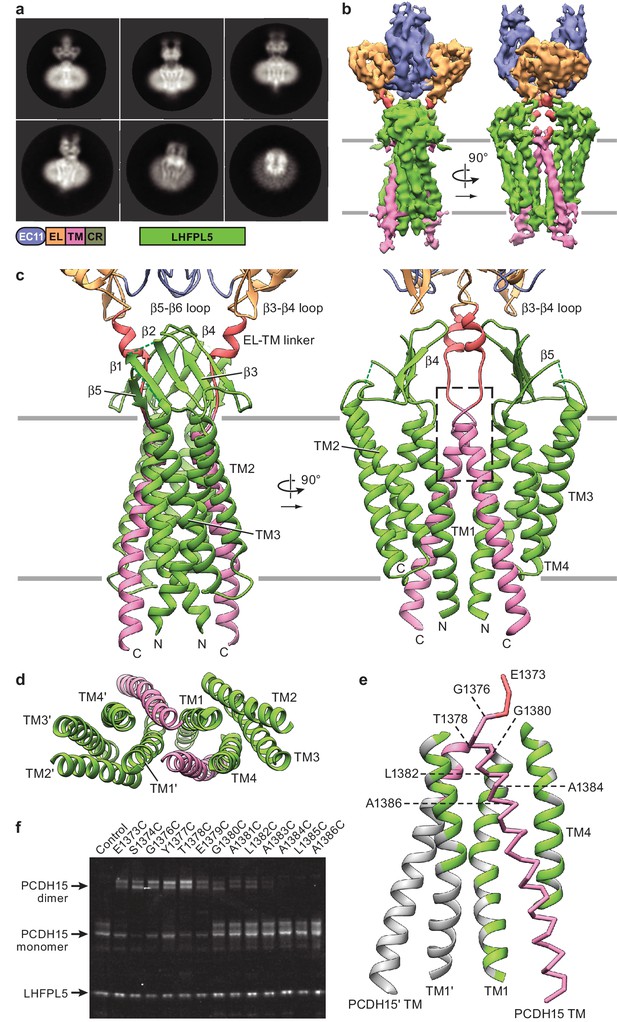
Cryo-EM structure of PCDH151EC/LHFPL5 complex.
(a) Representative 2D classes. Schematic of the corresponding constructs is shown. (b) 3D reconstruction. (c) Atomic model of the TM portion of the PCDH151EC /LHFPL5 complex. (d) View of PCDH15 and LHFPL5 TM helices from the extracellular side. (e) Interactions formed by the PCDH15 TM helix. One PCDH15 TM helix is shown as a backbone trace with interaction helices shown in cartoon representation, where only residues that potentially interact with the PCDH15 helix are colored. Residues mutated in panel (f) are indicated by labels. (f) SDS-PAGE analysis of site-directed cysteine crosslinking experiments. PCDH154EC Δ1413 and LHFPL5 in whole cell lysates expressing indicated mutants are detected using the fused fluorophores.
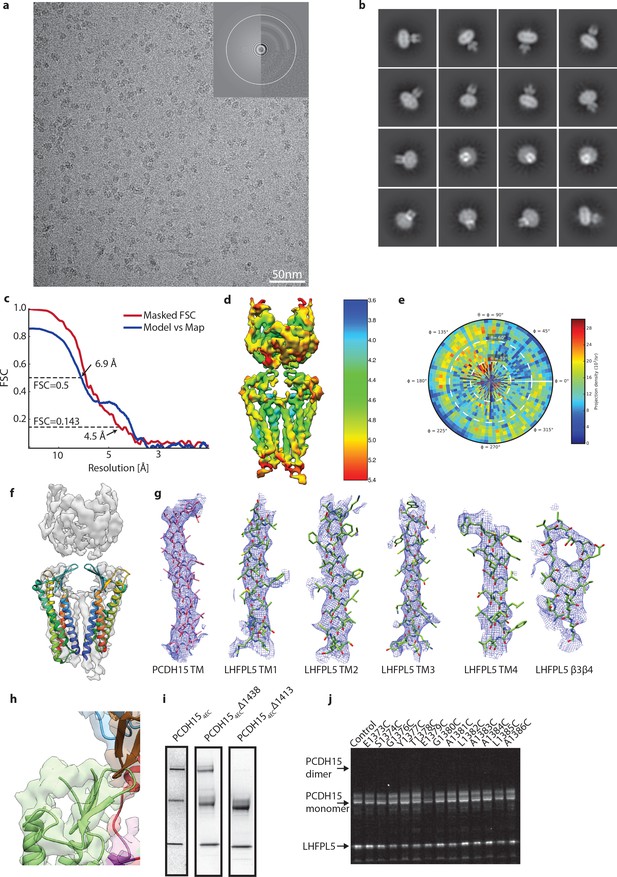
Details of PCDH151EC/LHFPL5 cryo-EM.
(a) Representative micrograph. (b) 2D class averages. (c) FSC analysis of final reconstruction. (d) Local resolution estimate. (e) Angular distribution. (f) Fit of claudin-15 crystal structure (PDB: 4P79) to cryo-EM density. (g) Local cryo-EM density around transmembrane helices and β3- β4 loop of LHFPL5. (h) Unaccounted density on top of PCDH15 β-sheet that should contain the TM3-β5 loop. (i) SDS-Page stained with coomassie of purified PCDH15/LHFPL5 complex using three different C-terminal truncations of PCDH15. Spontaneous dimer under nonreducing conditions is caused by residues 1411-1430, most likely C1414. (j) SDS-Page of PCDH154EC Δ1413 single-site cysteine mutants shown in Figure 4f under reducing conditions.
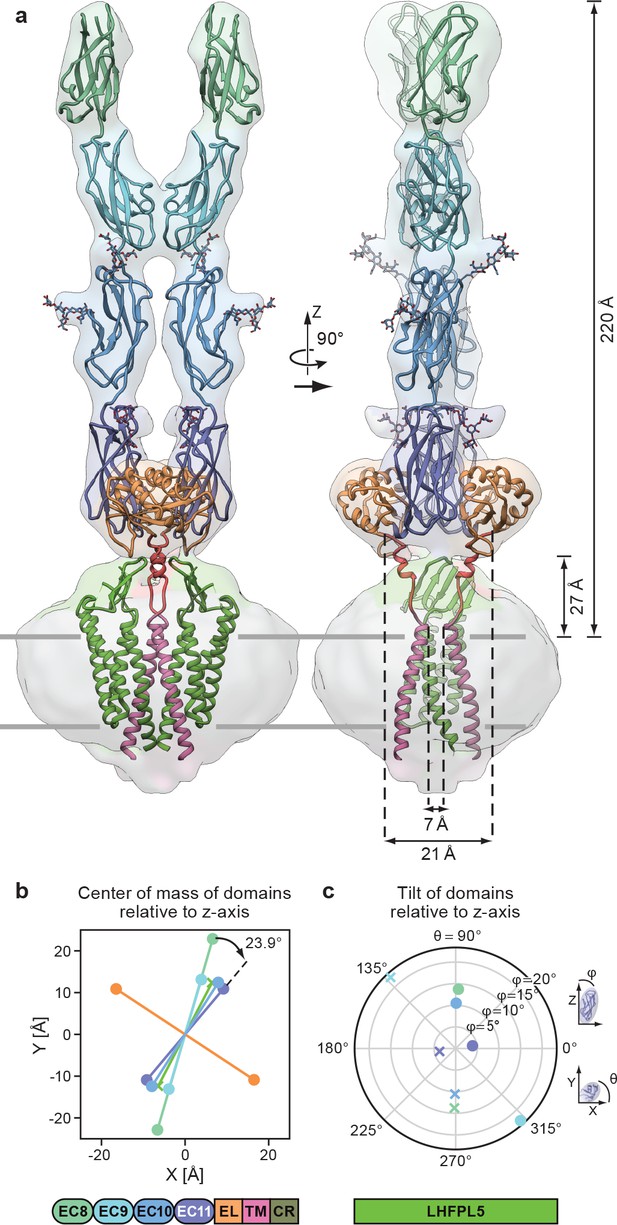
Complete model of the PCDH154EC/LHFPL5 complex.
(a) Complete model generated from fitting subunits into the PCDH154EC/LHFPL5 map, shown together with the map. Key dimensions are indicated. In the panel on the right hand side one LHFPL5 molecule has been omitted for clarity. (b) Center of mass of individual domains projected along the z axis, which is defined as the symmetry axis of the complex as indicated in panel a. A 23.9o twist of the EC11 compared to EC8 is indicated. (c) Tilt of the principal axes of the cadherin domain compared to the z axis. Domains of chains A and B are shown as dots and crosses, respectively. At the bottom of the figure is a schematic detailing the color coding of the individual domains.
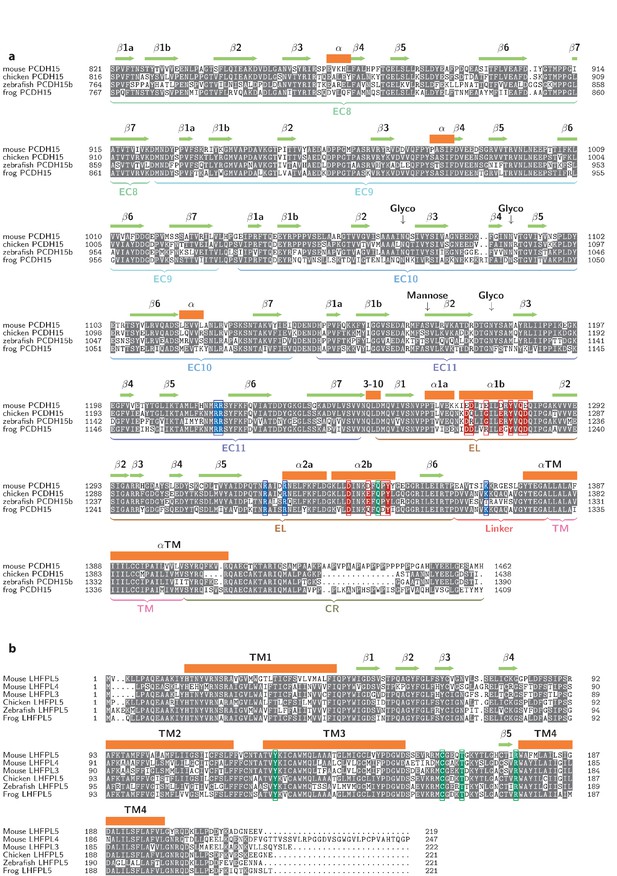
Multiple sequence alignments of PCDH154EC (a) and LHFPL5 (b).
Residues in the EC11-EL positive patch shown in blue. Residues in the EL negative patch shown in red. Residues, whose mutation cause disease, are shown in green.
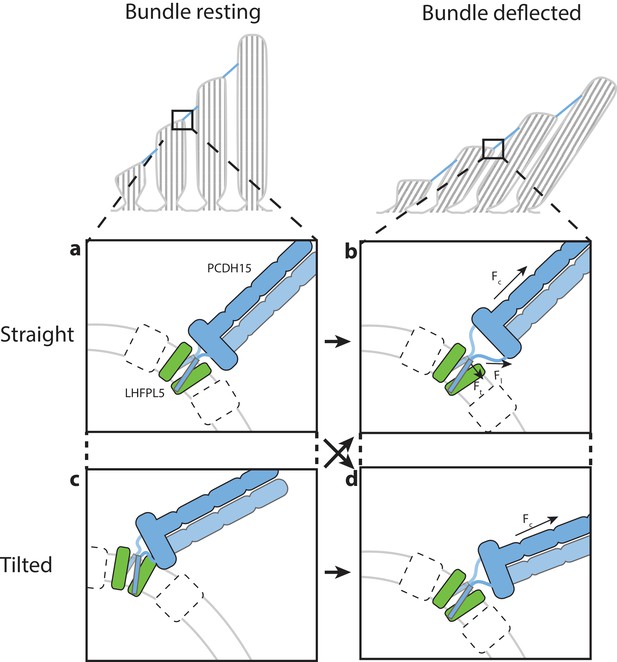
Hypothesis for the role of the PCDH154EC/LHFPL5 complex in mechanotransduction.
With the hair bundle of a hair-cell at rest the PCDH15 –LHFPL5 complex could either adopt the straight (a) or tilted-conformation (c), depending on whether the orientation of the membrane surrounding PCDH15 is perpendicular to the tip-link direction. Deflection of the hair bundle applies force to the PCDH15 cadherin domain chain (Fc) (b, d). This might result in the conversion from the tilted (c) to the straight conformation (b) or from the straight conformation (a) to the tilted conformation (d). In the case of the straight conformation (b) Fc is converted by the EC11-EL collar to a force on the linkers that, due to the local geometry, is no longer perpendicular to the membrane (Fl). We speculate that this could pull the PCDH15 transmembrane helices apart (Ft). White boxes with a dashed outline represent TMC, TMIE, or other so far unidentified proteins that could bind to either PCDH15 or LHFPL5 and sense this movement of the PCDH15 transmembrane helix or the change in membrane environment, ultimately leading to the opening of the MET channel.
Videos
Animation of 2D class averages of the PCDH154EC/LHFPL5 complex demonstrating the “split” conformation.
Animation of 2D class averages of the PCDH154EC/LHFPL5 complex demonstrating the “tilted” conformation.
Animation of 2D class averages of the PCDH154EC/LHFPL5 complex demonstrating the bending of the cadherin chain in the “bend” conformation.
Tables
Construct information.
https://doi.org/10.7554/eLife.38770.004| Construct name | Uniprot | Sequence range | N-terminal tag | C-terminal tag |
|---|---|---|---|---|
| PCDH154EC | Q99PJ1 | 1–30,821-1462 | None | mVenus-8xHis |
| PCDH151EC | Q99PJ1 | 1–30,1145-1462 | None | thrombin-mVenus-8xHis |
| PCDH15 EC11-ELCrys | Q99PJ1 | 1–30,1145-1380 | None | thrombin-mVenus-8xHis |
| PCDH15 EC11-ELAUC | Q99PJ1 | 1–30,1145-1380 | None | 8xHis |
| PCDH154EC Δ1438 | Q99PJ1 | 1–30,821-1438 | None | thrombin-mVenus-8xHis |
| PCDH154EC Δ1413 | Q99PJ1 | 1–30,821-1413 | None | thrombin-mVenus-8xHis |
| LHFPL5 | Q4KL25 | 2–219 | Strep-mCerulean-thrombin | None |
| TMIE | Q8K467 | 1–153 | None | thrombin-mCerulean-Strep |
| TMC1 | Q8R4P5 | 2–757 | Strep-mCerulean-thrombin | None |
Cryo-EM data collection, refinement and validation statistics.
https://doi.org/10.7554/eLife.38770.008| PCDH154EC/LHFPL5 (EMDB-7327) (PDB 6C13) | PCDH151EC/LHFPL5 (EMDB-7328) (PDB 6C14) | |
|---|---|---|
| Data collection and processing | ||
| Microscope | Titan Krios with Volta phase plate | Titan Krios |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 27 | 74 |
| Defocus range (μm) | 0.3–1.3 | 0.7–2.2 |
| Pixel size (Å) | 1.72 | 1.04 |
| Symmetry imposed | C2 | C2 |
| Initial particle images (no.) | 288,273 | 972,563 |
| Final particle images (no.) | 16,733 | 78,792 |
| Map resolution (Å) FSC threshold | 11.3 0.143 | 4.5 0.143 |
| Refinement | ||
| Initial model used (PDB code) | 4XHZ | 5B2G, 4P79, 5GJV, 6C10 |
| Model resolution (Å) FSC threshold | 13.8 0.5 | 6.9 0.5 |
| Model resolution range (Å) | ||
| Model composition Non-hydrogen atoms Protein residues | 8505 850 | 6749 862 |
| R.m.s. deviations Bond lengths (Å) Bond angles (°) | 0.003 0.72 | 0.006 1.25 |
| Validation MolProbity score Clashscore Poor rotamers (%) | 1.31 2.32 0.23 | 1.72 5.29 0.27 |
| Ramachandran plot Favored (%) Allowed (%) Disallowed (%) | 95.7 4.1 0.2 | 93.3 6.7 0 |
X-ray Data collection and refinement statistics
https://doi.org/10.7554/eLife.38770.014| EC11-EL | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.98 |
| Resolution range (Å) | 54.23–1.40 (1.45–1.40) |
| Space group | P 43 21 2 |
| Unit cell | |
| a, b, c (Å) | 57.68, 57.68, 159.18 |
| Α, β, γ (°) | 90, 90, 90 |
| Total reflections | 522785 (35483) |
| Unique reflections | 53914 (5195) |
| Multiplicity | 9.7 (6.8) |
| Completeness (%) | 99.73 (98.04) |
| Mean I/sigma(I) | 25.05 (2.32) |
| Wilson B-factor | 24.1 |
| R-merge | 0.036 (0.52) |
| R-meas | 0.037 (0.57) |
| R-pim | 0.011 (0.21) |
| CC1/2 (%) | 100 (30.7) |
| CC* (%) | 100 (68.5) |
| Refinement | |
| Reflections used in refinement | 53914 (5195) |
| Reflections used for R-free | 2696 (260) |
| R-work (%) | 16.9 (33.3) |
| R-free (%) | 19.5 (34.7) |
| CC(work) (%) | 96.5 (57.5) |
| CC(free) (%) | 95.7 (65.0) |
| Number of non-hydrogen atoms | 2141 |
| macromolecules | 1938 |
| ligands | 11 |
| solvent | 192 |
| Protein residues | 244 |
| RMS(bonds) | 0.006 |
| RMS(angles) | 0.75 |
| Ramachandran favored (%) | 98.76 |
| Ramachandran allowed (%) | 1.24 |
| Ramachandran outliers (%) | 0 |
| Rotamer outliers (%) | 0 |
| Clashscore | 2.55 |
| Average B-factor | 39.62 |
| macromolecules | 39 |
| ligands | 68.95 |
| solvent | 44.16 |
-
*Values in parentheses are for highest-resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | PCDH15 | Synthetic | UniProt: Q99PJ1 | |
| Gene (Mus musculus) | LHFPL5 | Synthetic | UniProt: Q4KL25 | |
| Gene (Mus musculus) | TMIE | Synthetic | UniProt: Q8K467 | |
| Gene (Mus musculus) | TMC1 | Synthetic | UniProt: Q8R4P5 | |
| Cell line (Spodoptera frugiperda) | Sf9 | ThermoFisher | 12659017 | |
| Cell line (Homo sapiens) | HEK293 tsa 201 | ATCC | CRL- 11268 RRID:CVCL_1926 | |
| Cell line (Homo sapiens) | HEK293S GnTI- | ATCC | CRL- 3022 RRID:CVCL_A785 | |
| Recombinant DNA reagent | pEG BacMam | doi: 10.1038/nprot.2014.173 | ||
| Recombinant DNA reagent | Lipofectamine 2000 reagent | Invitrogen | 11668–027 | |
| Recombinant DNA reagent | Cellfectin II reagent | Invitrogen | 10362–100 | |
| Software, algorithm | Unblur | doi:10.7554/eLife.06980 | http://grigoriefflab.janelia.org/ unblur | |
| Software, algorithm | UCSF MOTIONCOR2 | doi:10.1038/nmeth.4193 | http://msg.ucsf.edu/em/software/ motioncor2.html | |
| Software, algorithm | Gctf | doi:10.1016/j.jsb.2015.11.003 | http://www.mrc-lmb.cam.ac.uk/ kzhang/ | |
| Software, algorithm | cryoSPARC | doi:10.1038/nmeth.4169 | https://cryosparc.com/ | |
| Software, algorithm | RELION-2 | doi: 10.1016/j.jsb.2012.09.006 | RRID:SCR_016274 | http://www2.mrc-lmb.cam.ac.uk/ relion |
| Software, algorithm | DoG-picker | doi: 10.1016/j.jsb.2009.01.004 | http://emg.nysbc.org/redmine/ projects/software/wiki/DoGpicker | |
| Software, algorithm | Gautomatch | http://www.mrc-lmb.cam.ac.uk/ kzhang/ | ||
| Software, algorithm | Bsoft | doi:10.1016/j.jsb.2006.06.006 | https://lsbr.niams.nih.gov/bsoft/ | |
| Software, algorithm | UCSF Chimera | doi:10.1002/jcc.20084 | RRID:SCR_004097 | https://www.cgl.ucsf.edu/ chimera |
| Software, algorithm | PHENIX | doi:10.1107/S0907444912001308 | RRID:SCR_014224 | https://www.phenix-online.org |
| Software, algorithm | COOT | doi:10.1107/S0907444904019158 | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/ personal/pemsley/coot |
| Software, algorithm | Rosetta-CM | doi:10.1016/j.str.2013.08.005 | RRID:SCR_015701 | https://www.rosettacommons.org/docs/ latest/application_documentation/ structure_prediction/RosettaCM |
| Software, algorithm | GlyProt | doi:10.1093/nar/gki385 | RRID:SCR_001560 | http://glycosciences.de/modeling/ glyprot/php/main.php |
| Software, algorithm | CHARMM-Gui | doi:10.1002/jcc.20945 | RRID:SCR_014892 | http://charmm-gui.org/ |
| Software, algorithm | NAMD | doi.org/10.1002/jcc.20289 | RRID:SCR_014894 | http://www.ks.uiuc.edu/ Research/namd/ |
| Software, algorithm | MDFF | doi:10.1016/j.str.2008.03.005 | http://www.ks.uiuc.edu/ Research/mdff/ | |
| Software, algorithm | PyMOL | Schrodinger LLC | RRID:SCR_000305 | http://www.pymol.org |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38770.021