Nerfin-1 represses transcriptional output of Hippo signaling in cell competition
Figures
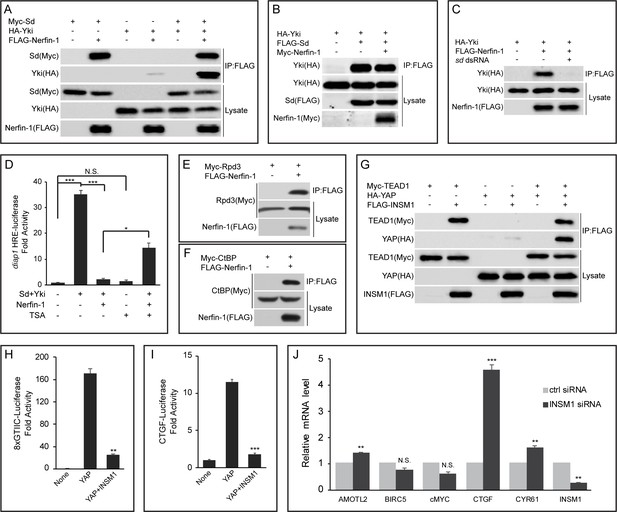
Nerfin-1 binds to Sd and antagonizes transcriptional activity of the Sd-Yki complex.
(A) S2R+ cells expressing the indicated constructs were subjected to IP by anti-FLAG. Note the strong interaction between FLAG-Nerfin-1 and Myc-Sd. Also note the weak interaction between FLAG-Nerfin-1 and HA-Yki, which was strengthened by expression of Sd. (B) The indicated constructs were transfected into S2R+ cells and subjected to IP by anti-FLAG. Note that Nerfin-1 did not affect Sd-Yki interaction. (C) The indicated constructs and dsRNAs were transfected into S2R+ cells and subjected to IP by anti-FLAG. Note that knockdown of endogenous sd abolished Nerfin-1-Yki interaction. (D) HRE-luciferase reporter was measured in triplicates in Drosophila S2R+ cells transfected with the indicated constructs, with or without the treatment of 200 nM TSA. Note that the activation of HRE-luciferase by Sd and Yki was suppressed by Nerfin-1, and this effect was reversed by TSA. The values are mean ± SEM, *p < 0.05, ***p < 0.001, N.S., non-significant. (E–F) Interaction between Nerfin-1 and Rpd3 (E) or CtBP (F). The indicated constructs were transfected into S2R+ cells and subjected to IP by anti-FLAG. (G) 293T cells expressing the indicated constructs were subjected to IP by anti-FLAG. Note the strong interaction between INSM1 and TEAD1. Also note the weak interaction between INSM1 and YAP, which was strengthened by expression of TEAD1. (H–I) 8xGTIIC-luciferase (H) or CTGF-luciferase (I) reporter was measured in triplicates in 293T cells transfected with the indicated constructs. The values are mean ± , **p < 0.01, ***p < 0.001. (J) Quantitative real-time PCR analysis of canonical YAP target genes from H727 cells transfected with control siRNA or siRNA against INSM1. Note the upregulation of AMOTL2, CTGF and CYR61. Transfection were performed in quadruplets and values are mean ± SEM, **p < 0.01, ***p < 0.001, N.S., non-significant.
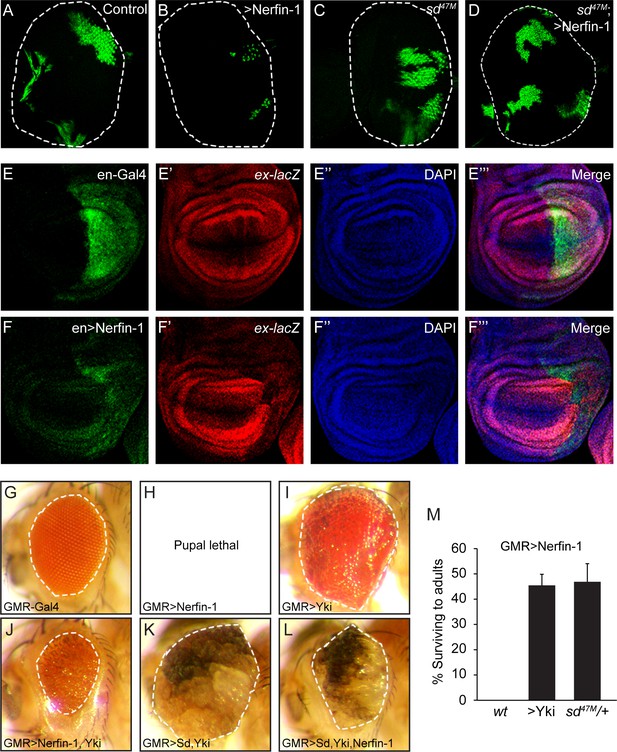
Genetic interactions between Nerfin-1, Sd and Yki.
(A–D) Eye discs containing GFP-marked MARCM clones of wildtype control (A), Nerfin-1 overexpression (B), sd47M mutant (C) or sd47M mutant with Nerfin-1 overexpression (D). Note the small size of Nerfin-1-overexpression clones and the normal size of sd47M mutant clones with Nerfin-1 overexpression. (E–F’’’) Wing discs expressing UAS-GFP only (E–E’’’) or UAS-GFP plus UAS-Nerfin-1 (F–F’’’) in the posterior compartment by the en-Gal4 driver. Note the reduced size of the posterior compartment and reduced expression of ex-lacZ upon Nerfin-1 overexpression (compare E-E’’’ and F-F’’’). (G–L) Adult eyes from flies overexpressing the indicated genes by the GMR-Gal4 driver, taken under the same magnification. (M) The percentage of GMR>Nerfin-1 flies surviving to adults was quantified relative to the expected number in the indicated genetic background (mean ± SEM). Three independent crosses were performed for each genotype. The complete genotypes are: (wt) GMR-Gal4/+; UAS-Nerfin-1/+; (>Yki) GMR-Gal4 UAS-Yki/+; UAS-Nerfin-1/+; (sd47M/+) sd47M/+; GMR-Gal4/+; UAS-Nerfin-1/+.
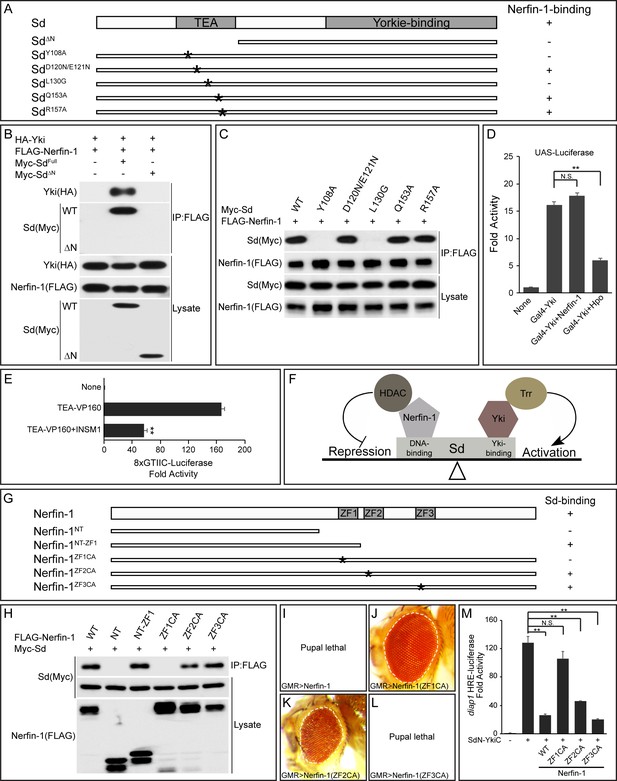
The TEA DNA-binding domain of Sd and the zinc fingers of Nerfin-1 are required for Sd-Nerfin-1 interaction.
(A) A summary of Sd mutants and their interactions with Nerfin-1. The domain structure of Sd and the location of the point mutations tested are also shown. (B–C) S2R+ cells expressing the indicated constructs were subjected to IP by anti-FLAG. Sd-Nerfin-1 interaction was abolished by deletion of the TEA domain (ΔN in B) or point mutations on the predicted hydrophobic surface of the TEA domain (Y108A and L130G in C), but not other point mutations in the TEA domain (D120N/E121N, Q153A and R157A in C). See Figure 3—figure supplement 1A–B for a detailed description of the point mutants tested. (D) UAS-luciferase reporter was measured in triplicates in S2R+ cells expressing the indicated constructs. Note the suppression of Gal4-Yki activity by Hpo, but not Nerfin-1. The values are mean ± SEM, **p < 0.01, N.S., non-significant. (E) 8xGTIIC-luciferase reporter was measured in triplicates in 293T cells expressing the indicated constructs. Note the repression of TEA-VP160 by INSM1. The values are mean ± SEM, **p < 0.01. (F) A model depicting the opposing activities of Nerfin-1 and Yki in regulating the transcriptional output of Sd, through the recruitment of enzymes conferring repressive (HDAC) and active (Trr) histone modification, respectively. (G) A summary of Nerfin-1 mutants and their interactions with Sd. The domain structure of Nerfin-1 and the location of the point mutations tested are also shown. (H) S2R+ cells expressing the indicated constructs were subjected to IP by anti-FLAG. Note that Sd-Nerfin-1 interaction was strongly abolished by mutation of ZF1 (ZF1CA), weakly abolished by mutation of ZF2 (ZF2CA), but not affected by mutation of ZF3 (ZF3CA). (I–L) Adult eyes from flies expressing the indicated Nerfin-1 mutants by the GMR-Gal4 driver. (M) HRE-luciferase reporter was measured in triplicates in S2R+ cells expressing the indicated constructs. The values are mean ± SEM, **p < 0.01, N.S., non-significant.
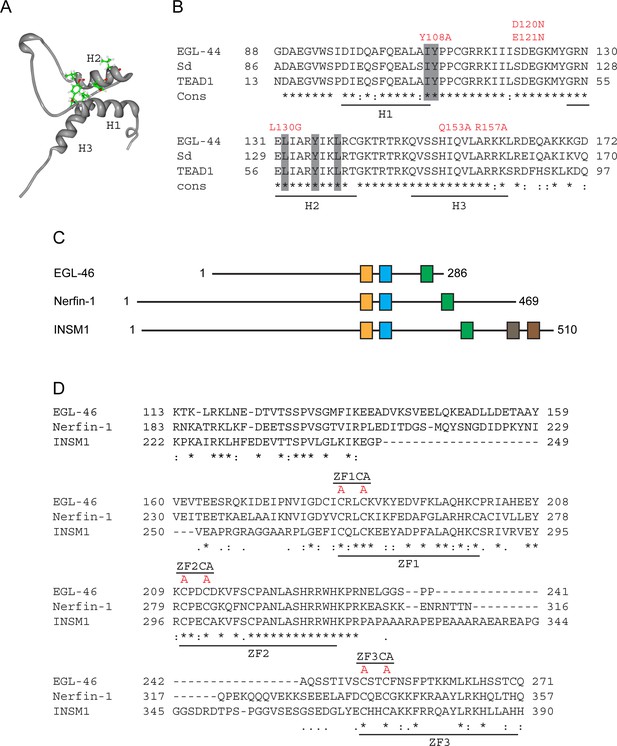
Structure-functional analysis of Sd and Nerfin-1.
(A) 3D structure of the TEA domain of human TEAD1 (Anbanandam et al., 2006). Side chains labeled in green represent hydrophobic residues in H1 and H2 that were hypothesized to form a protein-binding hydrophobic surface. H3 directly contacts DNA. (B) Sequence alignment of the TEA domain of C. elegans EGL-44, Drosophila Sd and human TEAD1. The three alpha-helices are underlined and the hydrophobic residues on the predicted protein-binding hydrophobic surface are highlighted by gray shading. Red letters indicate the point mutations tested for Sd-Nerfin-1 binding in Figure 3C. (C) A schematic diagram highlighting the ZFs of C. elegans EGL-46, Drosophila Nerfin-1 and human INSM1. The ZFs are colored differently to illustrate their relative conservation in different species. (D) Sequence alignment highlighting the ZFs of EGL-46, Nerfin-1 and INSM1. The ZFs are underlined. The red letters A indicate C-to-A mutations introduced into the respective ZF that were tested for Sd-binding and overexpression phenotype in Figure 3H–M.
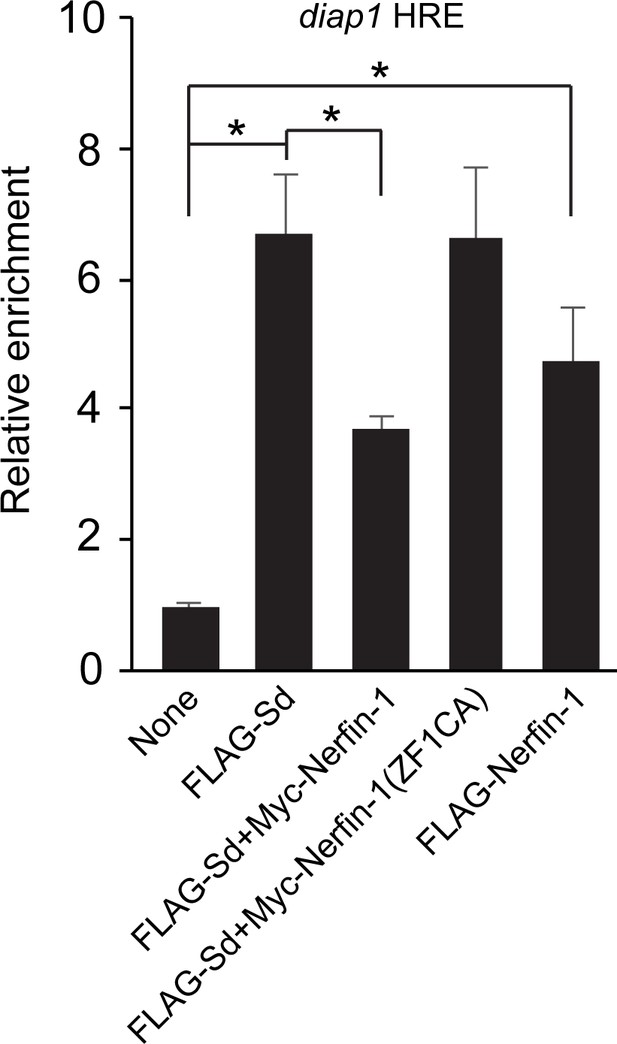
Nerfin-1 binds to the diap1 HRE site and impairs Sd-DNA binding.
Drosophila S2R+ cells expressing the indicated proteins were subjected to ChIP analysis using antibodies against FLAG. The enrichment of HRE at the endogenous diap1 locus was measured by real-time PCR. Both FLAG-Sd and FLAG-Nerfin-1 bound to the diap1 HRE. Note that Myc-Nerfin-1, but not Myc-Nerfin-1 (ZF1CA), impaired the binding of FLAG-Sd to the HRE site.
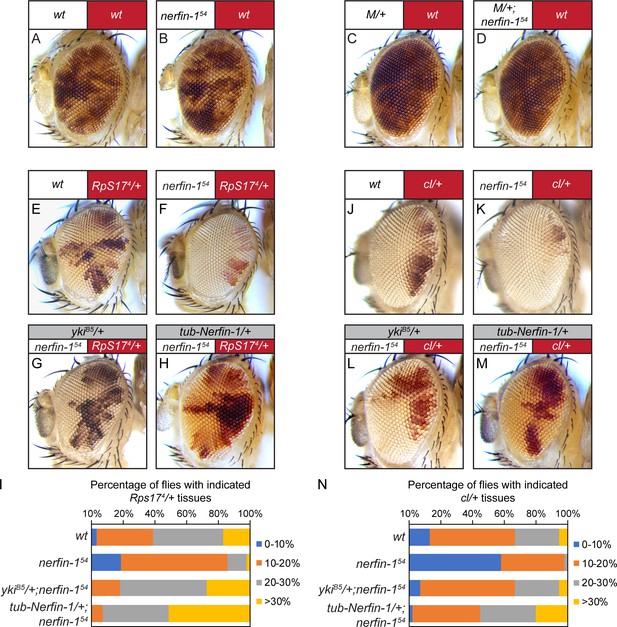
Loss-of-nerfin-1 in the winner cells enhances cell competition.
In all images of mosaic eyes, the genotypes of the constituting cells are indicated with matching colors (red or white) above the eye image. (A–B) Mosaic eyes containing control clones (A) or nerfin-154 mutant clones (B). Note the similar representation of nerfin-1 mutant clones compared to control clones. The complete genotypes are: (A) y w ey-Flp; FRT80B/P[w+] FRT80B; (B) y w ey-Flp; nerfin-154 FRT80B/P[w+] FRT80B. (C–D) Mosaic eyes containing Minute clones (C) or nerfin-154 mutant Minute clones (D). Note the similar representation of nerfin-1 mutant Minute clones compared to control Minute clones. The complete genotypes are: (C) y w ey-Flp/Df(1)R194 w; FRT80B/P[RpL36+w+] FRT80B; (D) y w ey-Flp/Df(1)R194 w; nerfin-154 FRT80B/P[RpL36+w+] FRT80B. See Figure 4—figure supplement 2E for a detailed description of the method to generate mosaic eyes with loss-of-nerfin-1 specifically in Minute cells. (E–I) Mosaic eyes of the indicated genotypes (E–H). The percentage of red-colored Minute tissue in each mosaic eye was measured and assigned to one of the following four groups: 0–10%, 10–20%, 20–30% and >30%. The number of flies in each group was counted and the percentage of each group was represented in bins in (I). The complete genotypes are: (E) y w ey-Flp; FRT80B/P[w+] Rps174 FRT80B (n = 105); (F) y w ey-Flp; nerfin-154 FRT80B/P[w+] Rps174 FRT80B (n = 101); (G) y w ey-Flp; ykiB5/+; nerfin-154 FRT80B/P[w+] Rps174 FRT80B (n = 105); (H) y w ey-Flp; tub-Nerfin-1/+; nerfin-154 FRT80B/P[w+] Rps174 FRT80B (n = 124). (J–N) Mosaic eyes of the indicated genotype (J–M). The percentage of red-colored tissue heterozygous for cell-lethal mutation in each mosaic eye was measured and assigned to one of the following four groups: 0–10%, 10–20%, 20–30% and >30%. The number of flies in each group was counted and the percentage of each group was represented in bins in (N). The complete genotypes are: (J) y w ey-Flp; FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 68); (K) y w ey-Flp; nerfin-154 FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 65); (L) y w ey-Flp; ykiB5/+; nerfin-154 FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 115); (M) y w ey-Flp; tub-Nerfin-1/+; nerfin-154 FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 151).

Nerfin-1 is dispensable for imaginal disc growth and follicle cell differentiation.
(A–D’) Pupal eye discs of the indicated genotypes were stained for DE-Cad to highlight cell outlines. In D-D’, a nerfin-1; nerfin-2 double mutant clone was marked as GFP negative. Note the similar number of interommatidial cells in each genotype. (E–F’) Wing discs containing GFP-negative nerfin-154 mutant clones, showing normal expression of Expanded (E–E’) or Merlin (F–F’) in the mutant clones. (G–G’’) A stage seven egg chambers containing GFP-negative nerfin-154 mutant follicle cell clones, showing the absence of Cut staining in the posterior follicle cells (marked by white dots). (H) Schematic diagram of the nerfin-2 locus, showing the intron/exon structure of the gene. Also shown are the two small deletions present in the nerfin-2m6-8 allele (see Materials and methods for nerfin-2 mutagenesis by CRISPR-Cas9). The first 7 bp deletion results in a frame shift of the coding sequence, introducing an early stop codon 20 bp downstream of the deletion.

Loss-of-nerfin-1 in the winner cells enhances cell competition.
(A–D) Mosaic eyes of the indicated genotype (A–C). The percentage of red-colored Minute tissue in each mosaic eye was measured and assigned to one of the following four groups: 0–10%, 10–20%, 20–30% and >30%. The number of flies in each group was counted and the percentage of each group was represented in bins in (D). The complete genotypes are: (A) y w ey-Flp; FRT80B/P[w+] RpL141 FRT80B (n = 49); (B) y w ey-Flp; nerfin-154 FRT80B/P[w+] RpL141 FRT80B (n = 47); (C) y w ey-Flp; ykiB5/+; nerfin-154 FRT80B/P[w+] RpL141 FRT80B (n = 36). (E) Diagram showing the crossing scheme for generating mosaic eyes with loss-of-nerfin-1 specifically in the loser cells. Df(1)R194 is a deficiency on the X chromosome that deletes the ribosomal protein-encoding gene RpL36. Males carrying ey-Flp on the X chromosome and nerfin-154 FRT80B on chromosomal arm 3L were mated to females carrying Df(1)R194 and a genomic DNA rescue construct (P[RpL36+w+] FRT80B) on 3L (Tyler et al., 2007). Mitotic recombination of 3L was induced in the eyes of the offspring by eyeless-Flp/FRT. Clones carrying Df(1)R194 and P[RpL36+w+] represented winner cells and were marked by red color, while twin spots carrying Df(1)R194 and nerfin-154 represented loser cells and were marked by white color. (F) A mosaic eye containing nerfin-154 tgi ΔP double mutant clones. Note the similar representation of double mutant clones (white-colored) compared to nerfin-154 mutant clones (Figure 4B). The complete genotype is: y w ey-Flp; nerfin-154 tgi ΔP FRT80B/P[w+] FRT80B. (G–H) The percentage of red-colored Minute tissue (G) or tissue heterozygous for cell-lethal mutation (H) in each mosaic eye was measured and assigned to one of the following four groups: 0–10%, 10–20%, 20–30% and >30%. The number of flies in each group was counted and the percentage of each group was represented in bins. Note the similar occupancy of Minute or cl/+ tissues in nerfin-154 and nerfin-154 tgi ΔP mosaic eyes. The complete genotypes are: (G) y w ey-Flp; FRT80B/P[w+] Rps174 FRT80B (n = 105), y w ey-Flp; nerfin-154 FRT80B/P[w+] Rps174 FRT80B (n = 101), y w ey-Flp; nerfin-154 tgi ΔP FRT80B/P[w+] Rps174 FRT80B (n = 115) and y w ey-Flp; tgi ΔP FRT80B/P[w+] Rps174 FRT80B (n = 92); (H) y w ey-Flp; FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 68), y w ey-Flp; nerfin-154 FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 65), y w ey-Flp; nerfin-154 tgi ΔP FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 89) and y w ey-Flp; tgi ΔP FRT80B/P[w+] l(3)CL-L1 FRT80B (n = 84).
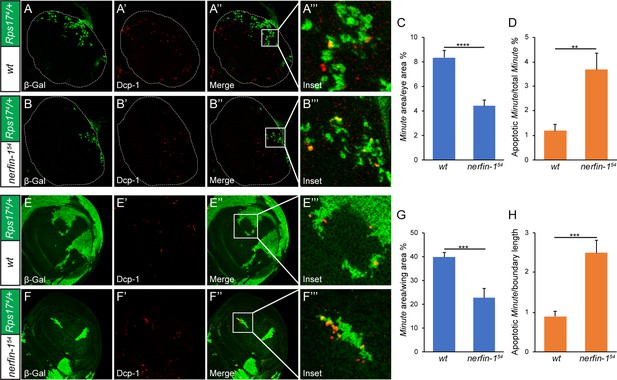
Developmental origin of the ‘super-winner’ phenotype conferred by loss-of-nerfin-1.
(A–B’’’) Mosaic larval eye discs of the indicated genotypes were stained for β-Gal (green; marking the Minute loser cells) and Dcp-1 (red). Enlarged views of the boxed area in (A’’) and (B’’) are shown in A’’’ and B’’’, respectively. The complete genotypes are: (A) y w ey-Flp; FRT80B/P[arm-lacZ w+] Rps174 FRT80B; (B) y w ey-Flp; nerfin-154 FRT80B/P[arm-lacZ w+] Rps174 FRT80B. (C) The percentage of Minute tissues in eye discs of the indicated genotypes in (A) and (B) was quantified (mean ± SEM, n = 15), ****p < 0.0001. Loss-of-nerfin-1 decreased the percentage of Minute tissues. (D) Quantification of apoptotic loser cells in eye discs of the indicated genotypes in (A) and (B). The number of cells positive for both Dcp-1 and β-Gal (indicating Minute cells undergoing apoptosis) relative to all β-Gal-positive cells (indicating all Minute cells) was plotted. The values are mean ± SEM, n = 15, **p < 0.01. (E–F’’’) Mosaic wing discs of the indicated genotypes were stained for β-Gal (green; marking the Minute loser cells) and Dcp-1 (red). Enlarged views of the boxed area in (E’’) and (F’’) are shown in E’’’ and F’’’, respectively. The complete genotypes are: (E) y w hs-Flp; FRT80B/P[arm-lacZ w+] Rps174 FRT80B; (F) y w hs-Flp; nerfin-154 FRT80B/P[arm-lacZ w+] Rps174 FRT80B. (G) The percentage of Minute tissues in wing discs of the indicated genotypes in (E) and (F) was quantified (mean ± SEM, n = 15), ***p < 0.001. (H) Quantification of apoptotic loser cells in wing discs of the indicated genotypes in (E) and (F). The number of cells positive for both Dcp-1 and β-Gal (indicating Minute cells undergoing apoptosis) was quantified per micron of boundary between the winner and loser cells, as described previously (Li and Baker, 2007). The values are mean ± SEM, n = 15, ***p < 0.001.
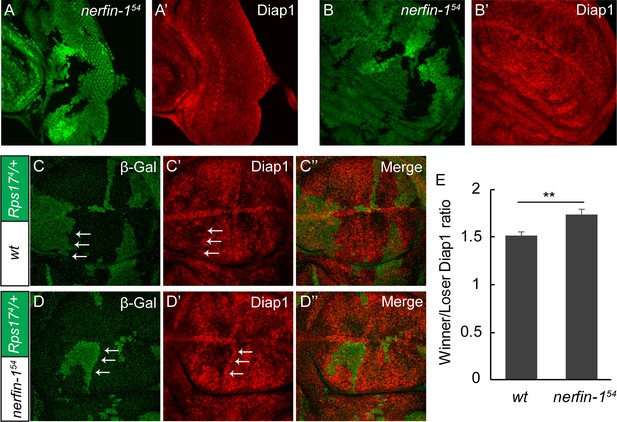
Nerfin-1 suppresses the expression of Yki target diap1 in the winner cells.
(A–B’) A third instar eye (A–A’) or wing disc (B–B’) containing GFP-negative nerfin-154 mutant clones was stained for Diap1 expression. Note the normal expression of Diap1 in mutant clones compared to neighboring wildtype cells. (C–E) Mosaic wing discs of the indicated genotypes were stained for β-Gal (green; marking the Minute loser cells) and Diap1 (red). The differential expression of Diap1 across the winner/loser boundary (a representative example of winner/loser boundary is marked by arrows) was quantified in (E) (mean ± SEM, n = 15), **p < 0.01. The complete genotypes are: (C) y w hs-Flp; FRT80B/P[arm-lacZ w+] Rps174 FRT80B; (D) y w hs-Flp; nerfin-154 FRT80B/P[arm-lacZ w+] Rps174 FRT80B.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38843.013





