Hair follicle epidermal stem cells define a niche for tactile sensation
Figures
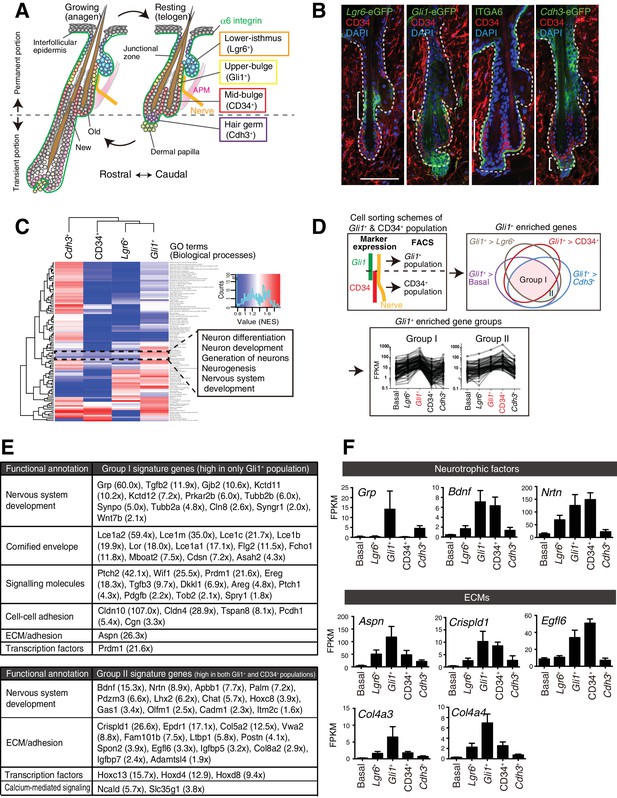
Upper-bulge epidermal stem cells are molecularly specialized for hair follicle–nerve interactions.
(A) Graphical illustration of epidermal stem cell compartments. APM, arrector pili muscle. (B) Distinct epidermal stem cell compartments in 8-week-old telogen skin were visualized with specific eGFP reporters and cell surface markers. Brackets indicate target cell populations for sorting. (C) Z-score heat map representing the normalized enrichment score (NES) of GSEA using the transcriptome data of each epidermal stem cell population. (D) Scheme of the extraction of genes highly expressed in the Gli1+ population (Group I) and in both the Gli1+ and CD34+ populations (Group II). (E) Lists of the genes highly expressed in Gli1+ population (Group I) and in both Gli1+ and CD34+ populations (Group II). (F) Expression levels of neurotrophic factor and ECM genes highly expressed in upper-bulge stem cells. Basal, basal epidermal stem cell pool. Data are mean ±SD, n = 3–4.
-
Figure 1—source data 1
Raw numerical data for Figure 1 and associated figure supplements.
- https://doi.org/10.7554/eLife.38883.004
-
Figure 1—source data 2
Normalized Enrichment Score (NES) of Gene Set.
- https://doi.org/10.7554/eLife.38883.005
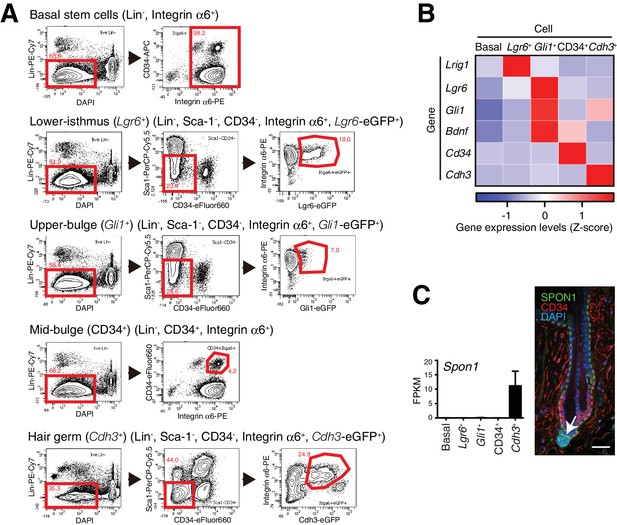
FACS-based cell isolation procedures for distinct epidermal stem cell populations.
(A) FACS sorting procedures of all basal epidermal cell populations using wild-type C57BL/6N mice, Lgr6+ lower-isthmus epidermal stem cells using Lgr6-GFP-ires-CreERT2 mice, Gli1+ upper-bulge epidermal stem cells using Gli1-eGFP mice, CD34+ mid-bulge epidermal stem cells using wild-type C57BL/6N mice, Cdh3+ hair germ epidermal cells using Cdh3-eGFP mice. Gates are indicated by red-line boxes and cells in the gates were further analysed in the next plots or sorted. The numbers in the plots represent the percentage of cells in the gates. Lin- indicates lineage-negative cells, which are negative for the markers of haematopoietic and endothelial cells (lineage-positive cells). (B) Z-score heat map representing qRT-PCR analysis of sorted cells with compartment-specific gene primers. See Methods for more detail. Data are mean of 3–4 independently isolated biological replicates. (C) Expression levels of Spon1 gene in different stem cell pools. Immunostaining pattern of SPON1 protein in 8-week-old telogen dorsal hair follicle was shown. White arrow indicates the restricted localization of SPON1 in dermal papilla and the basement membrane between dermal papilla and hair germ. This restricted expression and deposition of SPON1 corroborates little contamination of hair germ cells into the Gli1 + upper bulge population. FPKM, fragments per kilobase of transcript per million mapped reads. Basal, basal epidermal stem cell pool. Data are mean ± SD, n = 3–4. Scale bar, 10 μm.
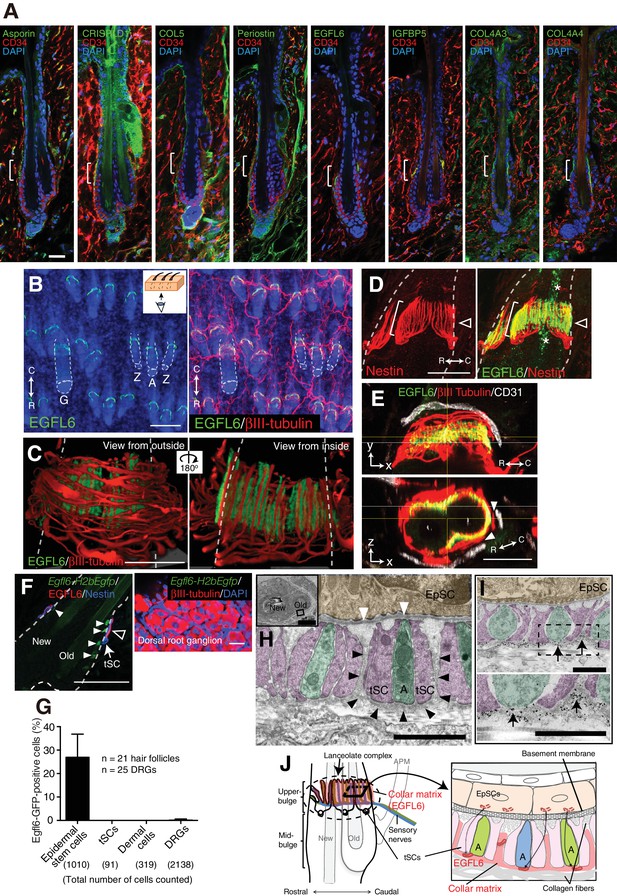
Upper-bulge epidermal stem cells deposit EGFL6 into the collar matrix.
(A) Immunostaining pattern of upper-bulge-specific ECM in 8-week-old telogen skin. Brackets: upper-bulge. (B) EGFL6 was colocalized with βIII-tubulin+ nerve endings in the upper-bulge of dorsal guard (‘G’), awl/auchene (‘A’) and zigzag (‘Z’) hair follicles. (C) Magnified images of EGFL6 and βIII-tubulin in telogen skin. (D) The protrusions of the terminal Schwann cells (white brackets) were colocalized with EGFL6 in the upper-bulge of 8-week-old telogen hair follicle (open arrowheads). Asterisks: nonspecific signals. (E) CD31+ blood vessels did not show direct contact with EGFL6 in 7-week-old telogen hair follicle. Arrowheads: gaps between EGFL6 (green) and blood vessels (white). (F) The dorsal skin and dorsal root ganglia (DRG) of an 8-week-old Egfl6-H2b-Egfp mouse was stained for eGFP, EGFL6, nestin and βIII-tubulin. eGFP (closed arrowheads) staining was observed in the upper-bulge (open arrowhead) epidermis of old bulge (Old), but not in other cellular components around the upper-bulge. (G) Statistical examination of Egfl6-H2BeGFP-positive cells around the upper bulge. tSC, termimal Schwann cell. Data are mean ± SD. (H) Transmission electron microscopic image of the upper-bulge of P35 zigzag hair follicle showing an electron-dense amorphous ECM structure, the collar matrix (black arrowheads) and ensheathed lanceolate complex endings [A, axon (green); tSC, terminal Schwann cell (pink); EpSC, epidermal stem cell (gold); white arrowheads, epidermal basement membrane]. (I) The EGFL6-gold particles were localized in the collar matrix (arrows). (J) Schematic representation of the localization of EGFL6 in the upper-bulge. Scale bars, 10 μm (A), 20 μm (C–F), 100 μm (B), 2 μm (H), 1 μm (I).
-
Figure 2—source data 1
Raw numerical data for Figure 2.
- https://doi.org/10.7554/eLife.38883.008
-
Figure 2—source data 2
List of ECM proteins screened for upper-bulge localization with immunohistochemical analysis.
- https://doi.org/10.7554/eLife.38883.009
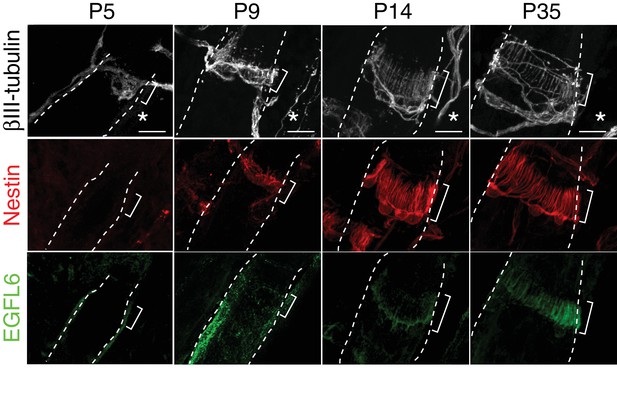
EGFL6 localization and lanceolate complex formation during hair follicle development.
Maximum projection images of P5, P9, P14 and P35 mouse dorsal hair follicles stained for βIII-tubulin, nestin and EGFL6. Brackets indicate upper-bulge regions. Asterisks indicate the caudal sides of dorsal skin. Scale bars, 10 μm.
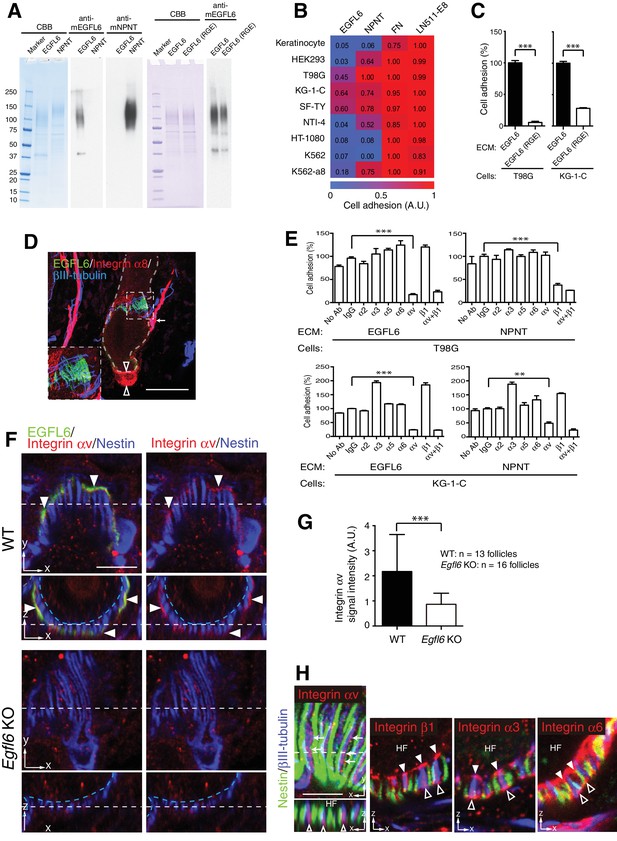
Cell adhesion to EGFL6 is mediated by αv integrins.
(A) Coomassie Brilliant Blue (CBB) staining and western blotting of purified EGFL6, RGE-mutant EGFL6, and nephronectin (NPNT) are shown. (B) Heat map display of the results of cell adhesion assays. The number of adhered cells was converted to arbitrary units (A.U.). FN, fibronectin; LN511-E8, laminin-511 E8 fragment. n = 3. Cell lines used are human primary keratinocytes, human embryonic kidney epithelial cell line HEK293, human glioblastoma cell line T98G, Human glioma cell line KG-1-C, human skin fibroblast cell line SF-TY, human embryonic fibroblast cell line NTI-4, human sarcoma cell line HT-1080, human myelogenous leukemia cell line K562 and K562 transfected with human integrin α8 cDNA K562-a8. (C) Cell adhesion assays with T98G and KG-1-C cells plated on EGFL6 or RGE-mutant EGFL6-coated dishes. Data are mean ± SEM, n = 3. (D) A maximum projection image of 8-week-old telogen dorsal hair follicles stained for EGFL6, integrin α8 and βIII-tubulin. Scale bar, 50 μm. Integrin α8 was detected in arrector pili muscles (white arrow), periphery of dermal papilla (open arrowheads) and periphery of lanceolate complexes (see the inset), but not in EGFL6-positive lanceolate complexes (see the inset). (E) Cell adhesion inhibition assays with integrin antibodies. Data are mean ± SEM, n = 3. (F) 8-week-old telogen dorsal hair follicles of wild-type and Egfl6 knockout mice were stained for EGFL6, integrin αv and nestin. Scale bar, 10 μm. Blue dashed lines indicate the surface of follicle epidermis and white dashed lines indicate cross-section positions. (G) Quantification of αv integrin signal intensity in lanceolate complexes. Data are mean ± SD. (H) 8-week-old telogen dorsal hair follicles of wild-type mice were stained for nestin, βIII-tubulin, integrins αv, β1, α3 and α6. Sagittal view (x–y) and transverse view (x–z) images are shown. HF, hair follicle. Scale bar, 5 μm. Statistics (C, E, G): two-tailed unpaired t-test.
-
Figure 3—source data 1
Raw numerical data for Figure 3.
- https://doi.org/10.7554/eLife.38883.011
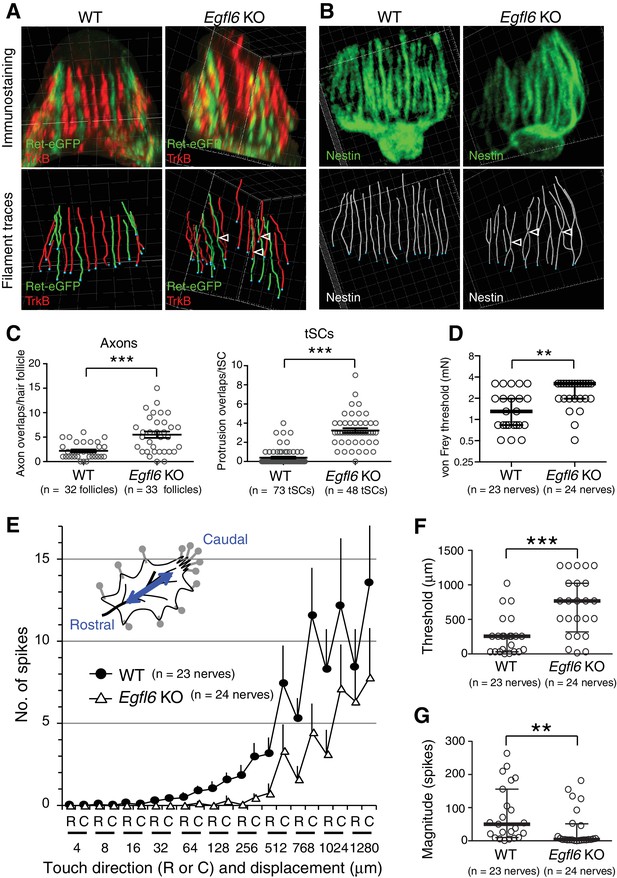
EGFL6 is required for the patterning and touch responses of lanceolate complexes.
(A and B) 3D reconstituted images and filament tracings of axonal endings (A) and terminal Schwann cells (B) in the old bulge of wild-type and Egfl6 knockout 8-week-old skin. Ret-eGFP, a marker for C-LTMRs and TrkB, a marker for Aδ-LTMRs. Open arrowhead: overlapping points of axonal endings and terminal Schwann cell processes. Grid width, 5 μm. (C) Overlapping points of axonal endings and terminal Schwann cell (tSC) processes were counted. Data are mean ± SEM. Two-tailed unpaired t-test. (D) Mechanical sensitivity of Aδ-LTMRs was analyzed using von Frey hairs in wild-type and Egfl6 knockout skin–nerve preparations from 10 to 12 week-old telogen skin. (E) Touch response patterns of Aδ-LTMRs in wild-type and Egfl6 knockout skin–nerve preparations. A ramp-and-hold touch stimulus was applied using a piezo-controlled micromanipulator (see Methods and Figure 4—figure supplement 5A). (F) Threshold displacement of Aδ-LTMRs. (G) Number of action potentials during the course of the touch stimulus protocol (i.e., magnitude of touch responses) was measured. Data are median with interquartile range (IQR) in (D, F, G) and mean ± SEM in (E). Statistics: Mann-Whitney U-test (D, F, G).
-
Figure 4—source data 1
Raw numerical data for Figure 4 and associated figure supplements.
- https://doi.org/10.7554/eLife.38883.018
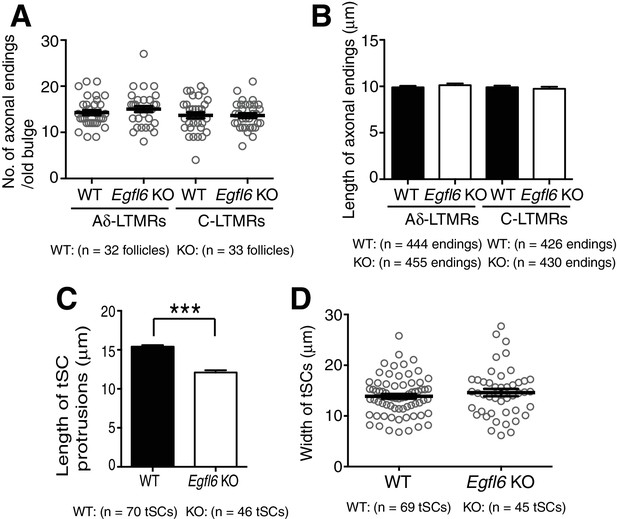
Quantification of morphological characteristics of axonal endings and terminal Schwann cell processes in wild-type and Egfl6 knockout mice.
(A) Number of axonal endings in the old bulge of zigzag hair follicles in 8-week-old mice was counted. Data are mean ± SEM, n = 32 hair follicles for wild-type and n = 33 hair follicles for knockout from three mice each. (B) Length of axonal endings in the old bulge of zigzag hair follicles was measured. Data are mean ± SEM, n = 444 axonal endings for wild-type and n = 455 axonal endings for knockout from three mice each. (C) Length and width of terminal Schwann cell processes were measured. Data are mean ± SEM, n = 70 tSCs (terminal Schwann cells) for wild-type and n = 46 terminal Schwann cells for knockout from three mice each. (D) Width of terminal Schwann cell processes were measured. Data are mean ± SEM, n = 69 terminal Schwann cells for wild-type and n = 45 terminal Schwann cells for knockout from three mice each.
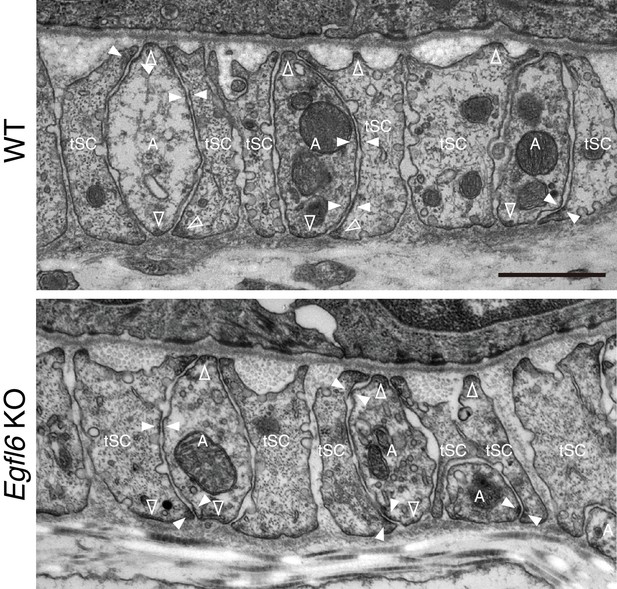
Ultrastructural examination of lanceolate complexes in wild-type and Egfl6 knockout mice.
Transmission electron microscopic images of the upper-bulge of P35 zigzag hair follicles of wild-type and Egfl6 knockout mice. Electron-dense plaques, characteristic structures for cell–ECM and cell–cell adhesions, are indicated by open arrowheads (cell–ECM adhesion sites) and closed arrowheads (cell–cell adhesion sites). A, axon; tSC, terminal Schwann cell. Scale bar, 1 μm.
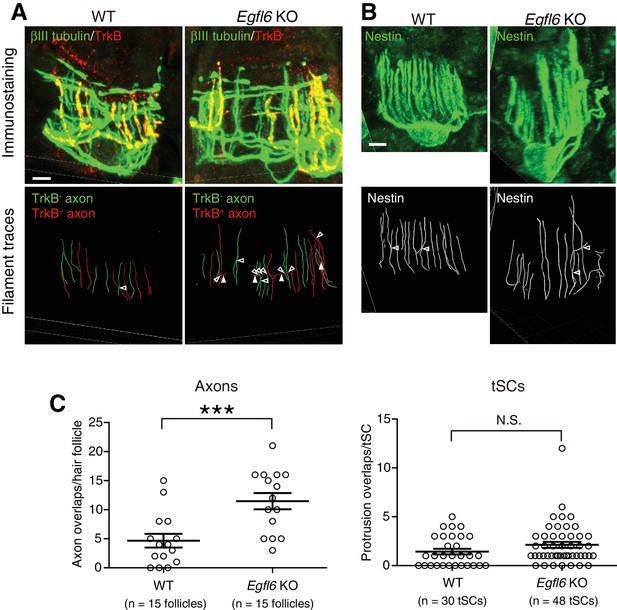
EGFL6 is required for the patterning of lanceolate complexes at the first telogen.
(A–B) 3D reconstituted images and filament tracings of axonal endings (A) and terminal Schwann cells (B) of zigzag hair follicles in wild-type and Egfl6 knockout mice at P19 first telogen. βIII-tubulin staining visualizes all LTMR axonal endings, including circumferential nerve endings. TrkB is a marker for Aδ-LTMRs. In zigzag hair follicles, TrkB- LTMRs are C-LTMRs. In the mutant, overlapping points of axonal endings (open arrowheads) were increased. This was partly due to the increase of horizontally extending branches of TrkB+ axonal endings (closed arrowheads) in Egfl6 knockout. These horizontally extending branches were also observed in wild-type lanceolate complexes with low frequency in the first telogen. Disorganized shape of terminal Schwann cells was occasionally observed in the mutant, but there is no significant difference in the number of terminal Schwann cell protrusion overlaps (see quantification below). Scale bars, 5 μm. (C) Overlapping points of axonal endings and terminal Schwann cell processes were counted. Data are mean ± SEM. Two-tailed unpaired t-test. Fifteen hair follicles from three wild-type mice and 15 hair follicles from three Egfl6 knockout mice were used for axon quantification. Thirty terminal Schwann cells from three wild-type mice and 48 terminal Schwann cells from three Egfl6 knockout mice were used for terminal Schwann cell quantification.
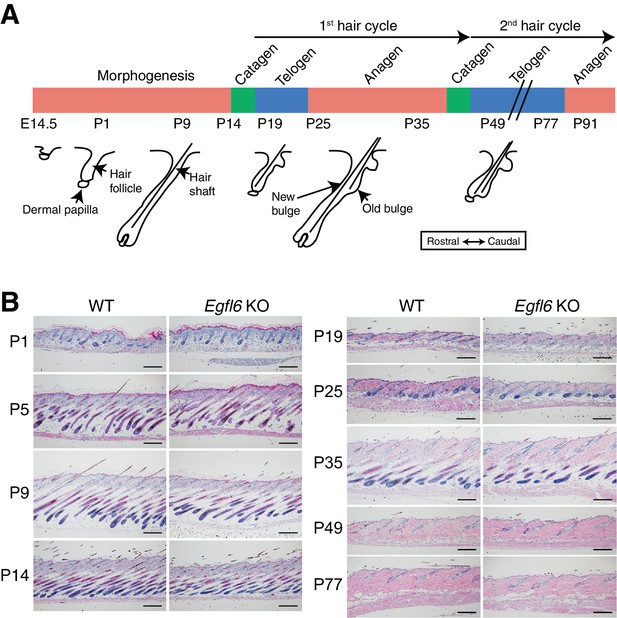
Histological examination of skin tissue morphology in wild-type and Egfl6 knockout mice.
(A) Schematic representation of mouse hair follicle morphogenesis and the following regeneration cycles. During the first anagen phase (P25–P35), old and new hair bulge structures form. (B) H&E-stained dorsal skin of wild-type and Egfl6 knockout mice. Scale bars, 100 μm.
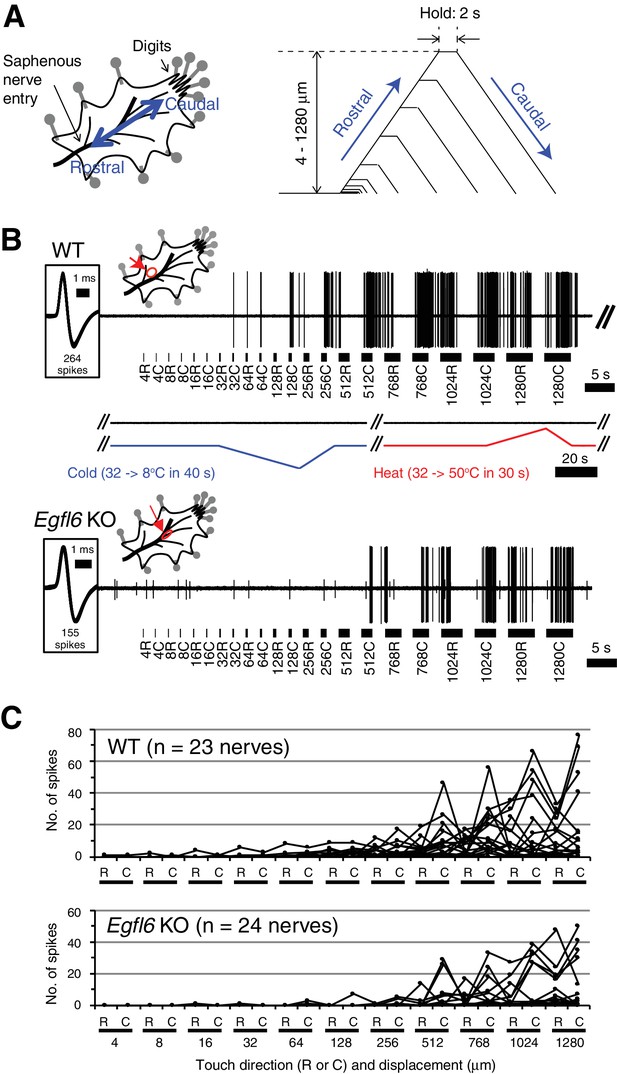
Skin–nerve preparation and recordings of responses of Aδ-LTMRs.
(A) Mouse skin–nerve preparation ex vivo, which is oriented with the dermis side up in the test chamber and affixed with pins. A ramp-and-hold touch stimulus protocol is shown. A probe tip was moved alternately in rostral and caudal directions with displacement in a progressively increasing manner (4–1280 μm) at a speed of 300 μm/s. The holding time was set for 2 s between the rostral and caudal movement of the tip. (B) Representative responses in the wild-type (upper panel) and Egfl6 knockout skin (lower panel). Note the first spike of the wild-type in response to touch stimulus with caudal displacement of 32 μm (i.e. 32C). No firings were observed in response to cold (from 32°C to 8°C in 40 s) or heat (from 32°C to 50°C in 30 s) stimulations. In the knockout, smaller responses to touch stimuli can be observed with the higher threshold displacement of 512 μm. Spike shapes are shown in insets squared at the left side. All of the spikes generated during a series of stimuli are superimposed (264 spikes in wild-type, and 155 spikes in knockout). The distribution of receptive fields is indicated by an arrow. (C) Individual response patterns in the wild-type (n = 23, upper panel) and the knockout (n = 24, lower panel).
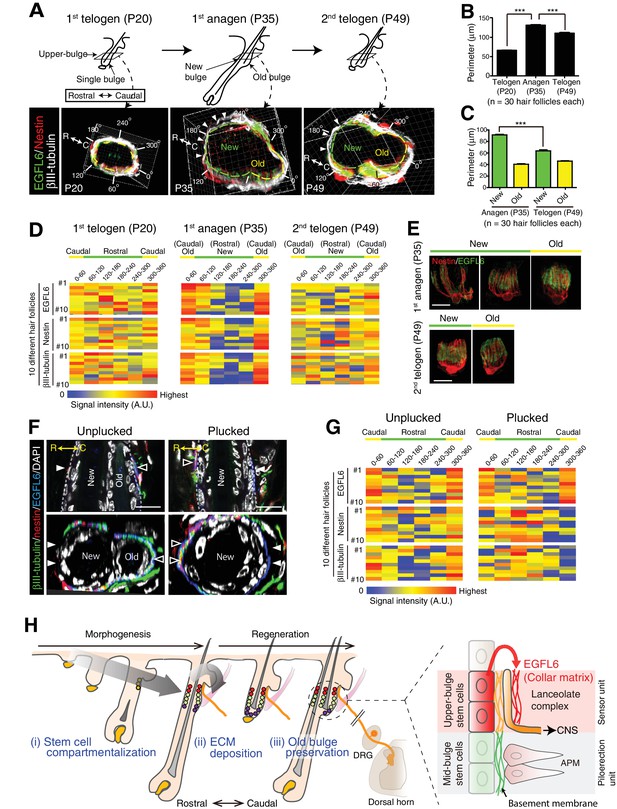
The old bulge provides stable epidermal–neuronal interfaces.
(A) Structural changes of the upper-bulge during the hair follicle regeneration cycle. Cross-sectional views of the upper-bulge of zigzag hair follicles are shown. Closed arrowheads indicate a large gap of lanceolate complexes. (B) Perimeter length of the upper-bulge and (C) the upper-bulge of new and old bulges. (D) Heat map of the relative signal intensity levels of EGFL6, nestin and βIII-tubulin along the upper-bulge perimeter. Geometry of the upper-bulge perimeter is shown above the heat map. (E) Morphological differences of EGFL6/terminal Schwann cell complexes in new and old bulges. (F) Sagittal and transverse sectional views of the upper-bulge in P35 anagen hair follicles with the club hair unplucked and plucked. Open and closed arrowheads indicate areas with and without lanceolate complexes, respectively. (G) Heat map of the relative signal intensity levels of EGFL6, nestin and βIII-tubulin along the upper-bulge perimeter in hair follicles with the club hair unplucked and plucked. (H) Schematic summary of the contribution of epidermal stem cells to sensory unit formation. (i) During development, epidermal stem cells for hair follicle–nerve interactions are induced and compartmentalized in the upper-bulge. (ii) Upper-bulge epidermal stem cells provide specialized ECM and neurogenetic environments for lanceolate complexes. (iii) epidermal stem cells maintain an old bulge at the caudal side of the hair follicle, providing stable hair follicle–nerve interfaces. Scale bars, 10 μm.
-
Figure 5—source data 1
Raw numerical data for Figure 5 and associated figure supplements.
- https://doi.org/10.7554/eLife.38883.024
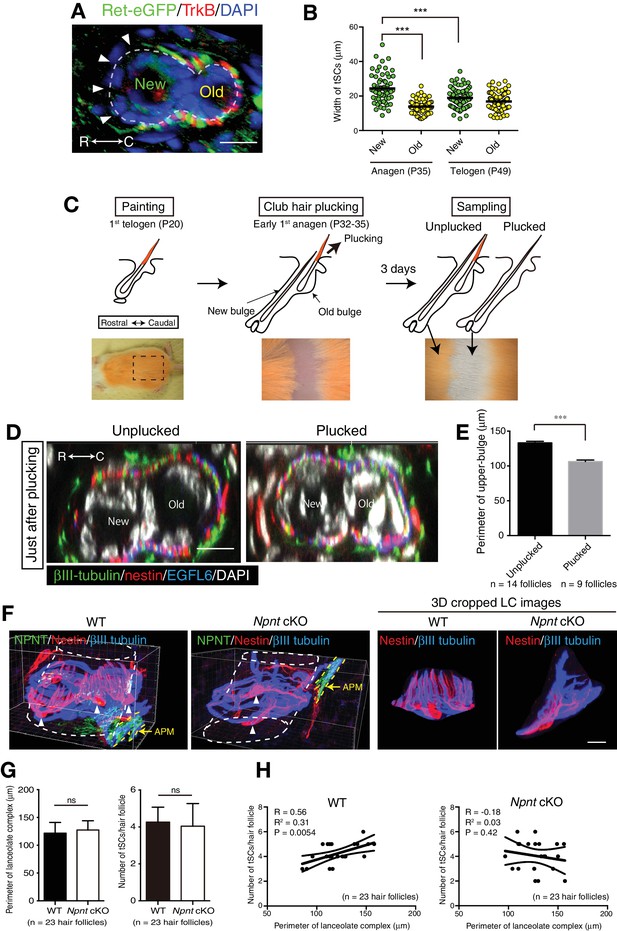
The old bulge formation provides stable epidermal-neuronal interfaces.
(A) A cross-sectional view of 7-week-old telogen upper-bulge stained for Ret-eGFP, TrkB and DAPI. Closed arrowheads indicate a gap of lanceolate endings. (B) The width of terminal Schwann cells in new and old bulges in the first anagen (P35) and second telogen (P49) follicles were quantified. n = 60–92 terminal Schwann cells from three mice. Individual measurement data are shown as circles. Mean ± SEM are shown as bars, n = 3 mice. Statistical comparisons were performed using two-tailed unpaired t-test. (C) Procedure of club hair plucking experiments. (D) Transverse sectional views of upper-bulge with club-hair unplucked and plucked P32 early anagen hair follicles stained for βIII-tubulin, nestin, EGFL6 and DAPI. Just after plucking, little structural damage to the old bulge epidermis and lanceolate complexes were observed. (E) Perimeter length of the upper-bulge of hair follicles 3 days after club hair plucking. n = 14 hair follicles from unplucked skin regions and nine hair follicles from plucked skin regions, respectively. (F) 3D reconstituted immunostaining images of upper-bulge region of 7-week-old telogen Awl/auchene hair follicles of wild-type and epidermis-specific conditional nephronectin (Npnt) knockout mice (Keratin5-Cre/Npnt fl/fl). Upon epidermal deletion of Npnt gene, arrector pili muscles (APM) moved above the bulge (Fujiwara et al., 2011), where lanceolate complexes were located. Arrowheads indicate the nuclei of terminal Schwann cells. In the mutant, some hair follicles showed disrupted patterning of longitudinal sensory nerve endings and terminal Schwann cells processes. This mutant image shows significantly fewer longitudinal lanceolate endings and terminal Schwann cell processes (see also 3D cropped lanceolate complex images). (G) The perimeter and number of lanceolate complexes in randomly selected non-guard hair follicles of wild-type and Npnt conditional knockout mice were measured. No statistically significant difference was observed. Data are mean ± SD, n = 23 hair follicles from wild-type and mutant mice, respectively. (H) The number of terminal Schwann cells was plotted against the perimeter of lanceolate complexes. In the wild-type, the number of terminal Schwann cells correlated well with the perimeter of lanceolate complexes, while in the Npnt knockout mice, distribution of the values of ‘number of tSC/perimeter of LCs’ was scattered and their correlation was lost (see R2 and p-value), indicating the anatomical changes of lanceolate complexes in Npnt mutants. tSCs, terminal Schwann cells. LCs, lanceolate complexes. n = 23 hair follicles from wild-type and mutant mice, respectively. Scale bars, 10 μm.
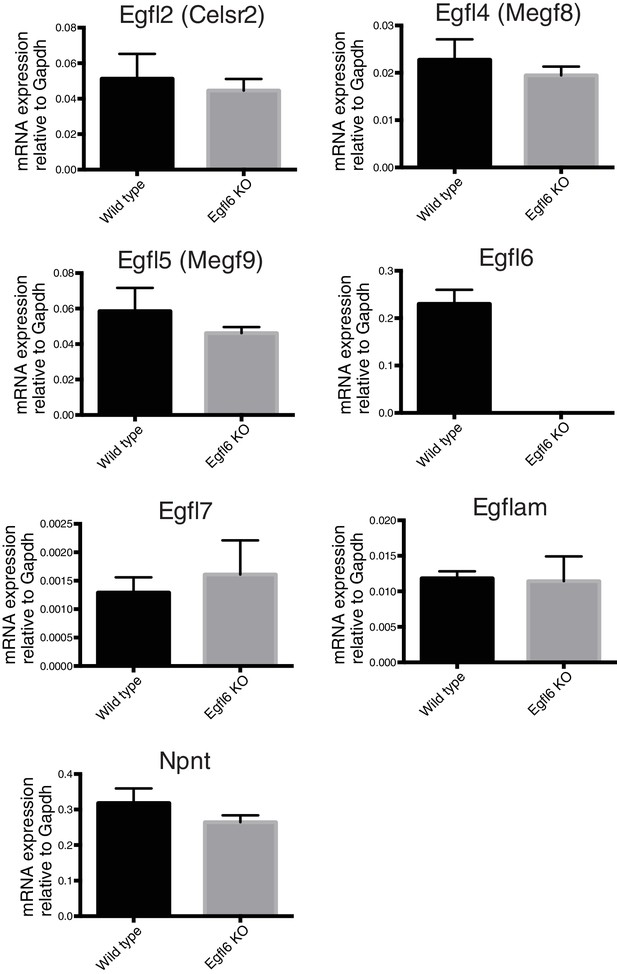
Expression of EGFL family genes in bulge epidermal stem cells of Egfl6 knockout mice.
CD34+epidermal stem cells, which contain mid-bulge epidermal stem cells and a part of Gli1+ upper-bulge epidermal stem cells, were isolated from 8-week-old wild-type and Egfl6 knockout mice. The expression of EGFL family genes were examined by qPCR. Data are mean ± SD, n = 3 mice for each genotype. The expression of 7 family members, including Egfl6, were detected in wild-type. But no significant difference in the expression of these genes was observed in the mutant (except for mutated Egfl6). The following genes were under detectable levels in both genotypes; Egfl3 (Megf6), Egfl8, Egfl9 (Dlk2), Megf10.
Tables
General characteristics of Aδ-LTMRs.
https://doi.org/10.7554/eLife.38883.019| Outcomes | WT (n = 23 nerves) | Egfl6 KO (n = 24 nerves) | P value |
|---|---|---|---|
| Activation threshold (V) | 3.0 (2.0–4.0) | 3.0 (2.0–6.0) | p=0.778 |
| Conduction velocity (m/s) | 6.6 (5.1–7.7) | 7.7 (5.0–8.6) | p=0.115 |
| Spontaneous activity (imp/s) | 0 (0–0) | 0 (0–0) | p>0.999 |
| Receptive field size (mm2) | 6.5 (4.7–13.2) | 4.8 (2.8–7.3) | p=0.059 |
-
Table 1–source data 1
Raw numerical data for Table 1.
- https://doi.org/10.7554/eLife.38883.020
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Egfl6 KO | MMRRC | 032277-UCD | |
| Genetic reagent (M. musculus) | Egfl6-H2b-Egfp BAC transgenic | this paper | ||
| Genetic reagent (M. musculus) | Ret-eGFP knock-in | PMID: 17065462 | Gift from H. Enomoto; Jain et al., 2006 | |
| Genetic reagent (M. musculus) | Egfl6 KO: Ret-eGFP knock-in | this paper | ||
| Genetic reagent (M. musculus) | Lgr6-GFP-ires-CreERT2 | Jackson Laboratories | 16934 | |
| Genetic reagent (M. musculus) | Gli1-eGFP | MMRRC | STOCK:Tg(Gli1-EGFP)DM197Gsat/Mmucd | |
| Genetic reagent (M. musculus) | Cdh3-eGFP | MMRRC | STOCK:Tg(Cdh3-EGFP)BK102Gsat/Mmnc | |
| Cell line (H. sapiens) | Primary keratinocytes | Watt Lab | Normal foreskin | |
| Cell line (H. sapiens) | HEK293 | RIKEN Bioresource Center | RCB1637 | |
| Cell line (H. sapiens) | T98G | JCRB Cell Bank | JCRB9041 | |
| Cell line (H. sapiens) | KG-1-C | JCRB Cell Bank | JCRB0236 | |
| Cell line (H. sapiens) | SF-TY | JCRB Cell Bank | JCRB0075 | |
| Cell line (H. sapiens) | NTI-4 | JCRB Cell Bank | JCRB0220 | |
| Cell line (H. sapiens) | HT-1080 | JCRB Cell Bank | JCRB9113 | |
| Cell line (H. sapiens) | K562 | JCRB Cell Bank | JCRB0019 | |
| Cell line (H. sapiens) | K562-a8 | PMID: 11470831 | Gift from L. Reichardt; Chavis and Westbrook, 2001 | |
| Cell line (H. sapiens) | 293F | Thermo Fisher | R79007 | |
| Antibody | Rabbit anti-mouse EGFL6 | this paper | CUK-1203–022 | Rabbit polyclonal; against aa 257–550; WB (0.2–2 μg/ml), IHC (1 μg/ml) |
| Antibody | Rabbit anti-mouse nephronectin | this paper | CUK-1192–006 | Rabbit polyclonal; against recombinant full-length mouse nephronectin-His protein; WB (0.4–2 μg/ml), IHC (2 μg/ml) |
| Antibody | Mouse anti-keratin 15 | Santa Cruz Biotechnology | LHK15 (sc-47697) | IHC (1:100) |
| Antibody | Mouse anti-bIII-tubulin | Abcam | 2G10 (ab78078) | IHC (1:250) |
| Antibody | Chicken anti-nestin | Aves Labs | NES0407 | IHC (1:250) |
| Antibody | Rat anti-nestin | Millipore | rat-401 (MAB353) | IHC (1:250) |
| Antibody | Rat anti-mouse CD31 | BioLegend | MEC13.3 | IHC (1:250) |
| Antibody | Chicken anti-GFP | Abcam | ab13970 | IHC (1:2000) |
| Antibody | Rabbit anti-GFP | MBL | 598 | IHC (1:1000) |
| Antibody | Rat anti-GFP | Nakarai | GF090R | IHC (1:75) |
| Antibody | Mouse anti-laminin γ1 | Millipore | MAB1914 | IHC (1:500) |
| Antibody | Rat anti-integrin αv | Abcam | RMV-7 | IHC (1:500) |
| Antibody | Hamster anti-mouse integrin αv | BD | H9.2H8 | IHC (1:250) |
| Antibody | Goat anti-human integrin αv | R and D Systems | AF1219 | IHC (1:100) |
| Antibody | Goat anti-mouse integrin α3 | R and D Systems | AF2787 | IHC (1:100) |
| Antibody | Hamster anti-mouse integrin β1 | BioLegend | HMb1-1 | IHC (1:100) |
| Antibody | Goat anti mouse integrin α8 | R and D Systems | AF4076 | IHC (2.5 μg/ml) |
| Antibody | Goat anti-TrkB | R and D Systems | AF1494 | IHC (4 μg/ml) |
| Antibody | Rat anti-CD45-PE-Cy7 | eBioscience | 30-F11 | FACS (1:100) |
| Antibody | Rat anti-TER-119-PE-Cy7 | eBioscience | TER119 | FACS (1:100) |
| Antibody | Rat anti-CD31-PE-Cy7 | eBioscience | 390 | FACS (1:100) |
| Antibody | Rat anti-Sca-1-PerCP-Cy5.5 | eBioscience | D7 | FACS (1:100) |
| Antibody | Rat anti-CD34- eFluor660 | eBioscience | RAM34 | FACS (1:100) |
| Antibody | Rat anti-CD49f-PE | eBioscience | GoH3 | FACS (1:100) |
| Antibody | Mouse anti-integrin α2 | DSHB | P1E6 | Cell adhesion inhibition (10 μg/ml) |
| Antibody | Mouse anti-integrin α3 | DSHB | P1B5 | Cell adhesion inhibition (10 μg/ml) |
| Antibody | Mouse anti-integrin α5 | DSHB | P1D6 | Cell adhesion inhibition (10 μg/ml) |
| Antibody | Rat anti-integrin α6 | BioLegend | GoH3 (313614) | IHC (1:500), Cell adhesion inhibition (10 μg/ml) |
| Antibody | Mouse anti-integrin αv | ATCC | L230 | Cell adhesion inhibition (10 μg/ml) |
| Antibody | Mouse anti-integrin β1 | DSHB | P5D2 | Cell adhesion inhibition (10 μg/ml) |
| Recombinant DNA reagent | BAC clone containing mouse Egfl6 genomic sequence | CHORI BACPAC Resources Center | RP23-124O13 | |
| Sequence-based reagent | qRT-PCR primers | This paper | See Table 2 | |
| Peptide, recombinant protein | EGF | Peprotech | 100 15 | |
| Peptide, recombinant protein | Insulin | Sigma | I5500 | |
| Peptide, recombinant protein | FLAG-tagged mouse full-length EGFL6 | this paper | ||
| Peptide, recombinant protein | FLAG-tagged mouse full-length EGFL6 (RGE mutant) | this paper | ||
| Peptide, recombinant protein | FLAG-tagged mouse full-length nephronectin | PMID: 21335239 | Fujiwara et al., 2011 | |
| Peptide, recombinant protein | Human plasma fibronectin | Wako | 063–05591 | |
| Peptide, recombinant protein | Laminin-511-E8 | Nippi | 892011 | |
| Commercial assay or kit | TruSeq Stranded mRNA Sample Prep Kit | Illumina | RS-122–2101 | |
| Commercial assay or kit | HiSeq SR Rapid Cluster Kit v2 | Illumina | GD-402–4002 | |
| Commercial assay or kit | HiSeq Rapid SBS Kit v2 | Illumina | FC-402–4022 | |
| Chemical compound, drug | 0.25% trypsin solution | Nakalai | 35555–54 | |
| Chemical compound, drug | Collagenase type I | Gibco | 17100–017 | |
| Chemical compound, drug | Hydrocortisone | Sigma | H4001 | |
| Chemical compound, drug | Cholera toxin | Sigma | C8052 | |
| Software, algorithm | HiSeq Control Software v2.2.58 | Illumina | http://www.illumina.com/systems/hiseq_2500_1500/software/hiseq-built-in-software.html | |
| Software, algorithm | RTA v1.18.64 | Illumina | http://support.illumina.com/sequencing/sequencing_software/real-time_analysis_rta.html | |
| Software, algorithm | bcl2fastq v1.8.4 | Illumina | http://jp.support .illumina.com/downloads/bcl2fastq_conversion_software_184.html | |
| Software, algorithm | FastQC v0.11.3 | Babraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | |
| Software, algorithm | FASTX Toolkit v0.0.14 | Hannon Lab | http://hannonlab.cshl.edu/fastx_toolkit/download.html | |
| Software, algorithm | Trim_galore v0.3.3 | Babraham Bioinformatics | http://www .bioinformatics.babraham.ac.uk/projects/trim_galore/ | |
| Software, algorithm | TopHat v2.0.14 | Johns Hopkins University | https://ccb.jhu. edu/software/tophat/index.shtml | |
| Software, algorithm | Cufflinks package v2.2.1 | Trapnell Lab | http://cole-trapnell-lab.github.io/cufflinks/ | |
| Software, algorithm | GSEA software 2.2.2 | Broad Institute | http://software. broadinstitute.org/gsea/index.jsp | |
| Software, algorithm | Bioconductor R with heatmap.2 gplots package | Gregory R. Warnes | https://www. rdocumentation.org/packages/gplots/versions/3.0.1/topics/heatmap.2 | |
| Software, algorithm | Imaris 7.2.1 | Bitplane | http://www.bitplane.com/imaris/imaris | |
| Software, algorithm | Volocity 6.3 | Perkin Elmer | http://cellularimaging.perkinelmer.com/downloads/ | |
| Software, algorithm | FilamentTracer | Bitplane | http://www.bitplane.com/imaris/filamenttracer | |
| Software, algorithm | DAPSYS data acquisition system | DAPSYS | http://www.dapsys.net/ | |
| Other | Piezo-controlled micromanipulator Nanomoter | Kleindiek Nanotechnik | MM3A | |
| Other | Feedback-controlled Peltier device | Intercross, Co. Ltd. | intercross-2000N | |
| Other | RNA-seq Fastq files | this paper | BioProject: PRJNA342736 | https://www.ncbi.n lm.nih.gov/bioproject/ |
Primer sequences for qRT-PCR.
https://doi.org/10.7554/eLife.38883.025| Target genes | Primer sequences |
|---|---|
| Gapdh (Forward) | TGCCAGCCTCGTCCCGTAG |
| Gapdh (Reverse) | CGGCCTTGACTGTGCCGTTG |
| Lrig1 (Forward) | TGTGTGTCTGAGAGACCCGAGC |
| Lrig1 (Reverse) | CAGAGCCACTGTGTGCTGTTGT |
| Lgr6 (Forward) | GGCTCCGAATCCTGGAGCTGT |
| Lgr6 (Reverse) | CTCAGGGTGGATGGCACGGA |
| Gli1 (Forward) | ATCTCCGGGCGGTTCCTACG |
| Gli1 (Reverse) | TGACTTCAGCTGGCAGGTTGC |
| Bdnf (Forward) | GCGGCGCCCATGAAAGAAGT |
| Bdnf (Reverse) | GGCCGAACCTTCTGGTCCTCA |
| Cd34 (Forward) | TGGCGCTGGGTAGCTCTCTG |
| Cd34 (Reverse) | TAGTCTCTGAGATGGCTGGTGTGG |
| Cdh3 (Forward) | TGGGGAAAGTAGCCTTGGCTGG |
| Cdh3 (Reverse) | CAGCCTCTGAGGGAAGGGACC |
| Egfl2 (Forward) | CCTGCCTCTGTCTGGATGGC |
| Egfl2 (Reverse) | GAAGCTGCGTGTGGTCACCT |
| Egfl3 (Forward) | GGCTCGCACTGTGTTCAGGT |
| Egfl3 (Reverse) | GGCACGTTTTGCCATCACCC |
| Egfl4 (Forward) | AGCTCTGCTCGGAGACACCA |
| Egfl4 (Reverse) | ACCAACTGCTGCCACTGCAT |
| Egfl5 (Forward) | CCTCCCCCAGAGCATATGTGTA |
| Egfl5 (Reverse) | AGCCGGTGACACCCACTTTG |
| Egfl6 (Forward) | AATACCCATCCGTGCAGCCC |
| Egfl6 (Reverse) | TCATGGTGCGCTTGTGAGCC |
| Egfl7 (Forward) | ACTTGCGACGGACACAGAGC |
| Egfl7 (Reverse) | GCGGATGCAACTCCCTCCAT |
| Egfl8 (Forward) | TGAAGAGAAGAAGCCCTCAGAGTGA |
| Egfl8 (Reverse) | CGCACACAAGGTCAGGTAGGG |
| Egfl9 (Forward) | CCTGCCAGAGCGGATGACTG |
| Egfl9 (Reverse) | CTTTCCTGCCCAGCCACTGT |
| Egflam (Forward) | CGGCTCTGCGAAAATCAGGC |
| Egflam (Reverse) | AACCGGGACGTTGTGTGACC |
| Megf10 (Forward) | GCCACAACGGAGCCTTTTGC |
| Megf10 (Reverse) | CCGCAGGGCACTTCTGTTCA |
| Npnt (Forward) | TCGCGTCCTCGCTCTACCTG |
| Npnt (Reverse) | ACACAGCTTTGAGCTGGCACC |
RNA-seq read counts and mapping statistics.
https://doi.org/10.7554/eLife.38883.026| Raw Read Count | Trimmed Read Count | rDNA Read Count | Multi-Mapped Read Count | Uniquely mapped Read Count | |
|---|---|---|---|---|---|
| Mouse_Basal_ Cells_Rep1 | 11,757,494 | 11,278,061 | 148,700 | 170,077 | 10,456,332 |
| Mouse_Basal_ Cells_Rep2 | 11,245,045 | 10,668,752 | 73,729 | 134,777 | 10,051,376 |
| Mouse_Basal_ Cells_Rep3 | 11,313,239 | 10,747,103 | 108,596 | 146,602 | 10,089,518 |
| Mouse_Bulge_ StemCells_Rep1 | 11,735,034 | 11,181,377 | 64,614 | 183,557 | 10,520,864 |
| Mouse_Bulge_ StemCells_Rep2 | 11,917,049 | 11,515,851 | 54,154 | 175,176 | 10,889,932 |
| Mouse_Bulge_ StemCells_Rep3 | 13,417,100 | 12,895,113 | 88,655 | 218,495 | 12,090,872 |
| Mouse_Cdh3+_ Cells_Rep1 | 10,875,593 | 10,409,342 | 68,193 | 177,407 | 9,574,901 |
| Mouse_Cdh3+_ Cells_Rep2 | 11,565,616 | 10,993,093 | 65,001 | 185,981 | 10,079,620 |
| Mouse_Cdh3+_ Cells_Rep3 | 11,405,265 | 10,746,768 | 76,143 | 206,384 | 9,790,096 |
| Mouse_Cdh3+_ Cells_Rep4 | 11,759,881 | 11,186,543 | 85,902 | 191,154 | 10,274,371 |
| Mouse_Gli1+_ Cells_Rep1 | 12,063,488 | 11,478,537 | 57,630 | 178,646 | 10,751,521 |
| Mouse_Gli1+_ Cells_Rep2 | 19,762,234 | 18,804,713 | 178,685 | 303,848 | 17,446,776 |
| Mouse_Gli1+_ Cells_Rep3 | 19,543,117 | 18,568,695 | 122,017 | 287,214 | 17,378,904 |
| Mouse_Lgr6+_ Cells_Rep1 | 20,044,455 | 19,058,807 | 121,507 | 307,804 | 17,798,594 |
| Mouse_Lgr6+_ Cells_Rep2 | 19,756,354 | 18,736,071 | 197,458 | 298,652 | 17,386,391 |
| Mouse_Lgr6+_ Cells_Rep3 | 19,639,299 | 18,680,969 | 217,502 | 298,146 | 17,210,635 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38883.027





