Epitope resurfacing on dengue virus-like particle vaccine preparation to induce broad neutralizing antibody
Figures

Comparison of the prM junction cleavage efficiency among different DENV-2 virus-like particles (D2VLPs).
(A) Schematic drawing of the prM protein. The C-terminal of the prM protein contains an α-helical domain (MH) in the stem region, followed by two transmembrane domains (MT1 and MT2). Numbers refer to the position of the amino acids in the polyprotein starting at the first amino acid of prM according to DENV-2 (NP_056776). Single letter designations of amino acid sequence alignment of representative strains from different serocomplexes of flaviviruses at the prM junction site includes dengue virus serotypes 1 – 4 (DENV-1 to DENV-4), Japanese encephalitis virus (JEV), St. Louis encephalitis virus (SLEV), West Nile virus (WNV), tick-borne encephalitis virus (TBEV), yellow fever virus (YFV), cell-fusion agent virus (CFAV), Zika virus (ZKV), immature DENV-2 VLP (imD2VLP) and mature DENV-2 VLP (mD2VLP). Numbers with P in the beginning refer to the positions of the amino acid relative to the prM cleavage site in the proximal direction (without apostrophe) and the distal direction (with apostrophe). Protein sequences were aligned, with the key P1 and P4 positions within the furin cleavage sites highlighted. The arrow indicates the prM cleavage site. The amino acids in red indicate the residues different from DENV-2. The PiTou 2.0 furin cleavage prediction scores are shown on the right for each sequence and the higher scores indicate the higher efficiency of furin cleavage. The relative quantity of prM and E of the wild-type and mutant DENV-2 VLP with P1-8 replacement (from other flaviviruses as shown) was measured by ELISA using MAb 3H5 (specific to E domain III of DENV-2) and MAb 155 – 49 (specific to DENV prM). The relative prM-to-E ratios were calculated by absorbance for prM/absorbance for E protein with reference to imD2VLP, whose pr portion was set as 100% uncleaved, as shown on the right. ND: not determined. Data are presented as means from three representative ELISA experiments with two replicates. (B) Culture supernatants of mD2VLP and imD2VLPs were collected and purified after electroporation with the respective plasmids. Five micrograms of proteins were loaded onto a 12% non-reducing Tricine-SDS-PAGE. E, prM and M proteins were assayed by Western blot using mouse hyper-immune ascitic fluids (MHIAF, 1:2000), MAb 2H2 (0.5 µg/mL) and anti-M protein mouse sera (1:25), respectively. E and prM proteins were visualized with enhanced chemiluminesence (ECL); however, M protein was visualized by TMB substrate to avoid high background. The number below each blot shows the relative densitometric quantification of E, prM and M protein bands by Bio-1D software.
Comparison of prM cleavage among different DENV-2 virus-like particles (D2VLP).
(A) Culture supernatants of mD2VLP and imD2VLPs were collected and purified after electroporation with the respective plasmids. Five micrograms of proteins were loaded onto a 12% non-reducing Tricine-SDS-PAGE. E, prM and M proteins were assayed by Western blot using mouse hyper-immune ascitic fluids (MHIAF, 1:2000), MAb 2H2 (0.5 µg/mL) and anti-M protein mouse sera (1:25), respectively. E and prM proteins were visualized with enhanced chemiluminesence (ECL); however, M protein was visualized by TMB substrate to avoid high background. (B) Percentages of pr-peptide remaining on particles were measured by calculating the relative densitometric quantity of prM band by Bio-1D software.
Comparison of physical properties of D2VLP among different DENV-2 virus-like particles (D2VLP).
(A) Equilibrium banding profiles of wtD2VLP (left), imD2VLP (center) and mD2VLP (right) after rate-zonal centrifugation on a 5 – 25% linear sucrose density gradient at 25,000 rpm for 3 hr. Equivalent amounts (50 μl) of D2VLP from each fraction were subjected to antigen-capture ELISA. (B) Electron micrographs at 50,000-fold magnification of wtD2VLP (left), imD2VLP (center) and mD2VLP (right) stained with uranyl acetate from the peak fraction in (A). (C) The diameter of 200 randomly selected particles of wtD2VLP (left), imD2VLP (center) and mD2VLP (right) from representative electron micrographs as in (B) were determined by software Gatan Digital Micrograph. (D) Endoglycosidase treatment of purified wtD2VLP, imD2VLP and mD2VLP. Samples were treated with PNGase F or Endo H and compared with untreated controls by 10% non-denaturing SDS-PAGE and immunoblotting with DENV-2 mouse hyper-immune ascitic fluid (MHIAF).
The structure of mature form virus-like particles of dengue virus serotype 2 (mD2VLP).
(A) The reconstructed cryo-EM map of the DENV VLPs (left panel) were presented with the radial color-code indicated (left), the size of the particle is 31 nm. Fitting of atomic E, M surface protein (PDB: 3J27) into cryo-EM density map (right) showed that the VLP has 60 copies of E in a T = 1 arrangement. The density map was shown as a transparent volume rendering into which was fitted the backbone structures of the E ecto-domain. The domains I, II, and III were highlighted in red, yellow and blue, respectively. The cage indicated the icosahedral symmetry. (B) The cross section showed the fitted E:M:M:E heterotetramer (PDB: 3J27) into DENV virion (left) and into mD2VLP (right). The map density was in mesh presentation. The atomic model of E:M:M:E heterotetramer was showed in ribbon. Domain definition of dengue E was the same as the previous description, the transmembrane domain of E was colored as light blue, the M protein was in orange color. It was clear that the density of H-layer which is composed of E-H1 to E-H3 and M-H was more solid while the density in T-layer which contains E-T1, E-T2, M-T1 and M-T2 was weak in VLP than in virion. The residues at 398, 401 and 412 in E-H1 of JEV sequence which were proved to play important role in promoting extracellular secretion (Purdy and Chang, 2005) were shown as magenta spheres. The transmembrane, perimembrane helices and lipid bilayer were labelled, the critical measurements were also shown.
The cryo-EM images and 2D analysis of the particles.
(A) Cryo-EM images of purified mD2VLP showed spherical particles (boxed) among irregular or incomplete structures (arrows). Irregularly-shaped particles were eliminated through visual inspection, while the spherical particles were subjected to further image analyses. (B) The particle size analyses showed the size variation in the sample and that the major peaks were located at ~26 nm,~31 nm and ~36 nm diameter size classes which were therefore denoted by small (‘S’), medium (‘M’) and large (‘L’). Each of the ‘S’, ‘M’ and ‘L’ particles identified was labeled. (C) The 2D image analyses showed that the particles had two distinct layers and that the particles classes with diameters of ~31 nm had more solid features than others.
The EM images of immunogold-labeled mD2VLPs.
The E proteins were labeled with domain III-specific monoclonal antibody (MAb 32 6), and conjugated with 6 nm gold particles.
The cryo-EM structure of m2DVLP.
The reconstructed cryo-EM map of the mD2VLPs was shown on the left. An asymmetric unit was indicated. On the right, the closest half of the density map has been removed to reveal the hollow structure. The resolution of the final reconstruction was determined to be 13.1 Å using a gold standard resolution estimate at cutoff 0.143 (lower panel).
Solvent accessibility of dengue virus serotype two virus soluble envelope (sE) protein (A, left) The plot of difference in relative solvent-accessible surface area (Δ%SASA).
A positive value of %SASA meant the residue became more exposed when the particle assembly shifts from virion (3 copies of E dimers per icosahedral asymmetric unit) to VLP (1 copy of E dimers per icosahedral asymmetric unit). The black dash line indicated the Δ%SASA >0.2, which was defined as highly exposed residues in VLP comparing to virion. The high positive values which were focused in the peptide regions such as the fusion loop peptide (including amino acid (aa) residues ranging from 100 to 110), aa 169–170 at domain I, aa 222–226, aa 239 and aa 251–262 at domain II as well as A strand of domain III (aa 300–308), the cd loop of domain III (aa 342–348) and G strand (aa 386–388) around the 5-fold openings. The residues interacting with MAb 1A1D-230, including residues 305–312, 352, 364, 388 and 390; the residues interacting with MAb 2D2231, including residues 67–72, 99, 101–104, 113, 177–180, 225–227, 247, 328, 384–386 (Heavy chain); 148–149, 153–155, 291–293, 295, 298, 299, 307, 309–310, 325, 327, 362–363 (light chain) and the residues interacting with human MAb EDE antibodies (Rodenhuis-Zybert et al., 2011), including aa residues 67–74, 97–106, 148–159, 246–249 and 307–314 were indicated. (A, right) The high positive peaks (Δ%SASA >0.2), low positive peaks (Δ%SASA between 0 and 0.2) and negative peaks (Δ%SASA <0) in the plot were colored by dark red, deem red and grey in the E dimer surface rendering. The groove located within E-dimer interface was outlined. (B) The highly exposed residues (Δ%SASA >0.2) which were colored by dark red were shown in the surface rendered E-dimer. The residues in E interacting with MAb 1A1D-2, 2D22 and EDE were in cyan spheres showing that they were highly exposed on the m2DVLP surface. Importantly, the binding footprints of the three antibodies were highly overlapping with footprint of highly exposed residues in m2DVLP, and formed a neutralization sensitive patch on m2DVLPs. The areas of the interacting epitopes are circled by black lines. (C) The E protein forming the rafts in virion were shown in the left panel where the three individual E proteins in the asymmetric unit are labeled A, B, and C of the E proteins, in the neighboring asymmetric unit are labeled A’, B’, and C’. The icosahedral 2-fold E protein dimers (B and B’) in m2DVLP have moved apart from each other causing the groove (right). The aa 101 which was responsible for DM25-3 antibody binding were shown in orange spheres.
The E protein rafts in a virion (left) and mD2VLP (right).
The E protein forming the rafts in the virion were shown in the left panel where the three individual E proteins in the asymmetric unit were labeled A, B, and C and the E proteins in the neighboring asymmetric unit are labeled A’, B’, and C’. The icosahedral 2-fold E protein dimers (B and B’) of mD2VLP moved apart from each other causing the groove (dashed line, right).

The solvent accessibility analyses.
Solvent accessibility of the dengue particle soluble envelope (sE) protein amino acid (aa) residues 1–396 in DENV virion and mD2VLP (middle and top, respectively). The all-atom model of VLP was built using MODELLER (da Silva Voorham et al., 2012), and the all-atom model virion was built based on the cryo-EM structure of the mature dengue virus at 3.5 Å resolution (PDB ID: 3J27). The relative solvent-accessible surface area (SASA) of both models was calculated using the POPS program. The plot shown represents the difference in relative solvent-accessible surface area (Δ%SASA) (Bottom). A positive value of %SASA means the residue becomes more exposed in mD2VLP than in DENV VLP. Domains I, II and III are highlighted in red, yellow and blue, respectively. The residues interacting with antibodies 2D22, EDE and 1A1D-2 are shown.
The ‘neutralization-sensitive hotspot’ on mD2VLPs.
(Left) The highly exposed residues (Δ%SASA >0.2) which were colored by dark red color are shown on the surface rendered VLP. The E dimer is outlined by a black dash line, the symmetry and the domains are labeled. The E epitopes interacting with MAb 1A1D-2 (center left), 2D22 (center right) and EDE (right) are colored in dark red on the surface showing that they were highly exposed on the mD2VLP surface and were highly overlapping with areas of highly exposed residues in mD2VLP.
Binding avidities of a panel of anti-E monoclonal antibodies (Mabs) to three types of D2VLP with different percentage of pr-peptide on particle surface.
Binding curves for (A) eight group-reactive Mabs recognizing E-protein of all four major pathogenic flavivirus serocomplexe, (B) four subgroup-reactive Mabs recognizing more than one flavivirus serocomplex, (C) Two complex-reactive Mabs recognizing all four serotypes of DENV serocomplex viruses, or (D) sub-complex-reactive Mabs recognizing more than one serotypes of DENV, (E) four serotype-specific-reactive Mabs recognizing DENV-2 only, were determined by antigen-capture ELISA. Original immunogen raised for generating murine Mabs and different domain of E protein recognized by Mabs were summarized in (F). Equal quantity of three types D2VLP were first titrated against purified D2VLP before adding to the wells. Mabs were 2-fold serial diluted starting from 1:1000 dilution fold. Data are expressed as P/N value by dividing the OD450 value from each dilution of Mab by the OD450 value from the control COS-1 culture supernatant. Data are means (with standard deviations for binding curves) for duplicates from three representative experiments.
-
Figure 4—source data 1
source data for antibody mapping results in Figure 4.
- https://doi.org/10.7554/eLife.38970.014
Total antigen-specific IgG, neutralizing titers and proportion of anti-prM antibodies compared among three groups of mice immunized with wtD2VLP, imD2VLP or mD2VLP.
(A) D2VLPs were concentrated and purified from clarified supernatants. The total protein concentration of purified imD2VLP and mD2VLP were first determined by the Bradford assay and then subjected to antigen-capture ELISA using 2-fold serial dilutions. The standard curve was used to titrate both antigens as equal amounts for the subsequent assays. (B) The endpoint IgG titer of 12 week post-immunization mouse sera was measured by antigen-capture ELISA, using equal amounts of homologous and heterologous purified D2VLP antigens. All endpoint titers were log10 transformed and depicted as geometric means with 95% confidence intervals. (C) The neutralizing antibody titers at 50% antigen focus-reduction micro neutralization (FRμNT50) in Vero cells infected with DENV-1 to 4. (D) Binding reactivity of serial dilutions of anti-wtD2VLP (left), anti-imD2VLP (center), and anti-mD2VLP (right) mouse sera were analyzed by ELISA using equal amounts of wild-type (imD2VLP) and mutant imD2VLP (Δ2H2) antigens. (E) Proportions of 2H2-like, anti-prM antibodies from two different D2VLP immunization groups were calculated based on the formula 100*[(OD450imD2VLP-OD450Δ2H2)/OD450imD2VLP] at a 1:1000 dilution of mouse sera. All data presented are based on a representative of three independent experiments with two replicates from n = 5 mice sera per group per experiment and expressed as mean ±SEM. The statistical significance was determined using the two-tailed Mann-Whitney U test to account for non-normality of the transformed data. *p<0.05; **p<0.01; ****p<0.0001.
-
Figure 5—source data 1
source data for antibody mapping results in Figure 5.
- https://doi.org/10.7554/eLife.38970.020

Proportions of anti-prM antibodies from both D2VLP immunization groups were measured using an epitope-blocking ELISA.
Percent blocking of HRP-labeled anti-prM MAb (2H2-HRP) by sera from mice vaccinated with imD2VLP or mD2VLP was determined by the formula 100*[(OD450imD2VLP-OD450imD2VLP blocked by MAb 2H2)/OD450imD2VLP] using a 1:1000 dilution of mouse sera.
Epitope identification of MAbs 2H2 and 155–49.
Single (K26P), double (K26P and K21D) and triple (K26P and K21D and F1A) mutations were performed on imD2VLP-expression plasmids by site-directed mutagenesis. Various D2VLP mutants were expressed in COS-1 cells by electroporation with plasmid DNAs as indicated and tissue culture media were clarified 3 days after transfection for antigen-capture ELISA. Substitutions of K26P on imD2VLP led to a significantly reduced binding activity of MAb 2H2; however, a weak binding can still be detected for MAb 155–49. Double mutation didn’t affect the binding of MAb 155–49 on imD2VLP, compared to a single mutation. Only the triple mutation completely abolished the binding of MAb 155–49 on imD2VLP. The antigens used were standardized at a single concentration with an optical density (OD) of 0.8, which was based on the standard curves generated by antigen-capture ELISA. Data are shown as means ± SEM from three independent experiments. P/N ratio refers to the antibody binding magnitude between designated VLP-containing (P) and VLP-free culture supernatants (N) by dividing the absorbance of P by that of N.
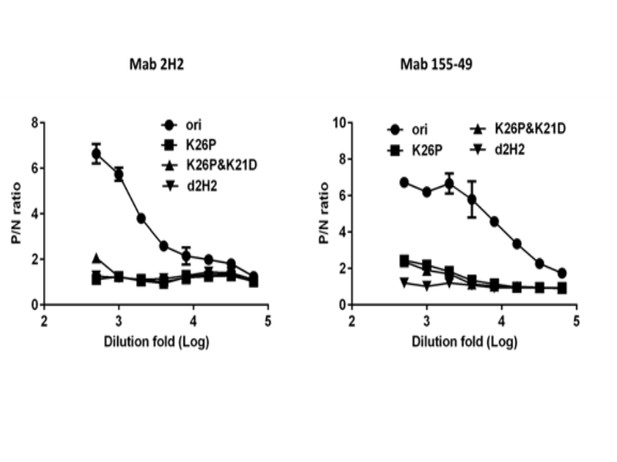
Serial dilutions of imD2VLP and mutant imD2VLP (Δ2H2), containing mutations at the MAb 2H2 binding site (F1A, K21D, K26P), were tested for binding with MAb 2H2 (left) and control antibody 3H5 (right) by ELISA.
P/N ratios refer to the antibody binding magnitude between designated VLP-containing (P) and VLP-free culture supernatant (N) by dividing the absorbance of P by that of N.

Binding ELISA was performed to test the reactivity of sera from mice immunized with two doses of imD2VLP or mD2VLP against equal amounts of imD2VLP and mD2VLP antigens.
The difference in binding activity was converted to a bar chart at 1:1000-fold dilutions of mice sera. Proportions of DM 25–3-like antibodies from two different D2VLP immunization groups were calculated based on the formula 100x(OD450mD2VLP-OD450imD2VLP)/OD450mD2VLP. The data are presented as means ± SEM from three independent experiments with two replicates. The two-tailed Mann-Whitney U test was used to test statistical significance. *p<0.05.
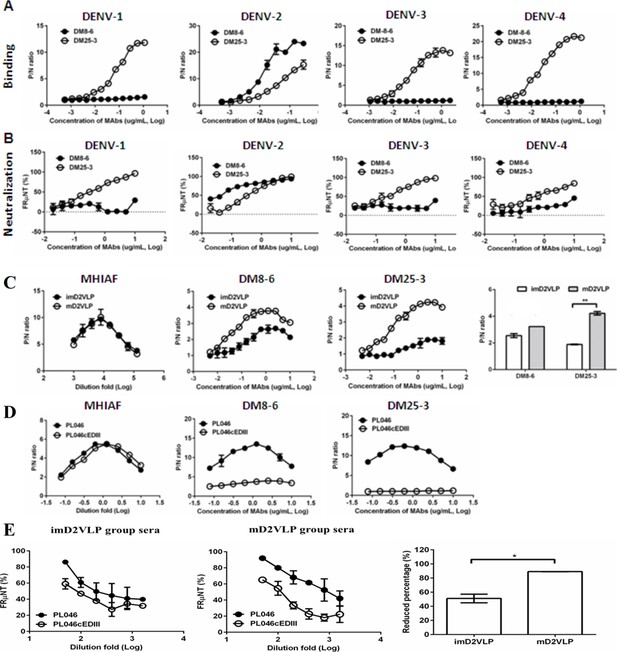
Characterization of murine monoclonal antibodies (MAbs) generated from mouse splenocytes following immunization with mD2VLP.
Binding (A) and neutralizing (B) activities of MAbs DM8-6 and DM25-3 against DENV-1 to 4 were measured by ELISA and the focus-reduction micro-neutralization test (FRμNT). (C) Binding curves of MAb DM8-6 (center left) and DM25-3 (center right) against imD2VLP and mD2VLP were performed using ELISAs. Equal amount of both imD2VLP and mD2VLP was properly titrated by antigen-capture ELISA using mouse hyper-immune ascitic fluid (MHIAF) against DENV-2 (left). The difference in binding activities of both MAbs is presented by a bar graph (right). (D) A recombinant DENV-2 virus was produced by replacing domain III with a consensus sequence of domain III (PL046cEDIII) (Fibriansah et al., 2015a) and the binding activity of DM8-6 (center) and DM25-3 (right) was compared with that of parental DENV-2 strain PL046. Equal amount of both PL046 and PL046cEDIII was properly titrated by antigen-capture ELISA using mouse hyper-immune ascitic fluid (MHIAF) against DENV-2 (left). (E) FRμNT of two-fold diluted mice sera immunized with mD2VLP and imD2VLP against parental PL046 and PL046cEDIII DENV-2 viruses (n = 5 per group per experiment). The differences in FRμNT50 from mD2VLP and imD2VLP immunization groups were converted to bar chart at 1:1000 fold dilution of mice sera. The conversion was based on the formula 100*[FRμNT50 of (PL046- PL046cEDIII)/FRμNT50 of PL046]. P/N ratio refer to the antibody binding magnitude between designated VLP-containing (P) and VLP-free culture supernatant (N) by dividing the absorbance of P by that of N. The data are presented as means ± SEM from three independent experiments with two replicates. The two-tailed Mann-Whitney U test was used to test statistical significance. *p<0.05. **p<0.01.
-
Figure 6—source data 1
source data for antibody mapping results in Figure 6.
- https://doi.org/10.7554/eLife.38970.025

Identification of the neutralizing epitopes of MAbs DM25-3 and DM8-6.
(A) Bar graph shows the decrease in MAb reactivity in a binding-ELISA for mD2VLPs with substitutions at the designated residues. Various mD2VLP mutants were expressed in COS-1 cells by electroporation with plasmid DNAs as indicated. Tissue culture media were harvested 3 days after transfection and clarified for antigen-capture ELISA. Substitutions at N103K, G104Q, K307E, E314R, T315H, P364R and W391G produced plasmids, which did not secrete measurable VLP antigens into the tissue culture medium. Therefore, only mutant plasmids indicated secreted VLP antigens and were used for epitope mapping by binding ELISA. (Top) Substitutions of E311E led to a significantly reduced binding activity of neutralizing MAb DM8-6, compared to mD2VLP. (Bottom) Substitutions of W101G led to a complete loss of binding by MAb DM25-3. Data shown are a representative experiment of three independent experiments. (B) To ensure that the antigenic structures were intact, mutant VLPs including W101G, K310E and E311R were further confirmed by using a panel of DENV-2 MAbs, including group-cross-reactive antibodies (4G2, 4A1B-9, 6B3B-3, 6B6C-1, DM25-3) recognizing all four major pathogenic flavivirus serocomplexes; complex cross-reactive antibodies (T5-1) recognizing all four DENV complex viruses, and type-specific antibody (DM8-6) recognizing DENV-2 only. The relative binding of MAbs was determined by dividing the OD450 of mD2VLP mutants by that of mD2VLP. The data are presented as means ± SEM from three independent experiments with two replicates. The two-tailed Mann-Whitney U test was used to test statistical significance.
Transmission electron microscope (TEM) examination of negatively stained W101G-mutant mD2VLP.
Purified W101G-mutant mD2VLP were adsorbed onto a glow-discharged carbon coated grid (EMS CF-200-Cu) for 1 min, and inspected by TEM. The images were taken by a JEM1400 electron transmission microscope at a magnification of 30,000x using a 4k × 4 k Gatan 895 CCD camera. The diameters of particles were between 30 and 32 nm as measured by ImageJ software.
Binding ELISA was performed to test the reactivity of sera from mice immunized with two doses of wtD2VLP, imD2VLP or mD2VLP against equal amounts of imD2VLP and fusion-peptide amino acid 101 mutant imD2VLP antigens (imD2VLP-W101G).
(A) Serial dilutions of imD2VLP and mutant imD2VLP (W101G), containing mutations at the MAb 4G2 binding site (amino acid residue 101 mutation from tryptophan (W) to glycine (G)), were tested for binding with MAb 4G2 (Top) and control antibody 3H5 (Bottom) by ELISA. (B) Equal amounts of imD2VLP and imD2VLP-W101G were used to test the binding activity of sera from three groups of mice receiving three different types of VLP antigens. P/N ratios refer to the antibody binding magnitude between designated VLP-containing (P) and VLP-free culture supernatant (N) by dividing the absorbance of P by that of N. (C) The difference in binding activity was converted to a bar chart at 1:1000-fold dilutions of mice sera. Proportions of 4G2-like antibodies from three different D2VLP immunization groups were calculated based on the formula 100x(OD450imD2VLP-OD450imD2VLPW101G)/OD450imD2VLP. The data are presented as means ± SEM from three independent experiments with two replicates. The two-tailed Mann-Whitney U test was used to test statistical significance. *p<0.05, **p<0.01.
Schematic presentation of the schedule and survival curves for mouse immunization and challenge.
(A) Groups of four 4-week-old, female, ICR mice were injected intramuscularly with imD2VLP and mD2VLP at week post vaccination (wpv) 0 and 4 at a dose of 4 μg/100 μL. Mice were bled from the retro-orbital sinus at week 4 following the second injection, and individual mouse serum collected from immunized females 1 week prior to mating was evaluated for the presence of the total IgG titer and the virus neutralization response by ELISA and focus-forming micro-neutralizing assay (FRNT). For the evaluation of passive protection by maternal antibody, ICR pups from the mating of non-immunized males with immunized females 11 weeks post initial vaccination were obtained for viral challenge. Pups from unvaccinated females were used as the challenge control. ICR pups from the designated groups were challenged individually through intracranial route at 2 days after birth with 104 focus-forming unit (FFU) which were equivalent 141, 61, 11, 1000 times of 50% lethal doses (LD50) of DENV-1, to DENV-4, respectively. The percent survival of the mice was evaluated daily for up to 21 days. (B) Survival curve of pups delivered from the female mice receiving mD2VLP, imD2VLP monovalent vaccine or TNE control, then challenge with DENV-1 to 4 after birth. The in vivo protective efficacy of DENV-2 monovalent vaccine is maturity-dependent. N in parentheses indicated the numbers of pups of each group. Kaplan-Meier survival curves were analyzed by the log-rank test. * p<0.05, **p<0.01.
-
Figure 7—source data 1
source data for antibody mapping results in Figure 7.
- https://doi.org/10.7554/eLife.38970.027
Additional files
-
Supplementary file 1
Nucleotide sequences of primers for site-directed mutagenesis used in this study
- https://doi.org/10.7554/eLife.38970.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38970.029





