Differing isoforms of the cobalamin binding photoreceptor AerR oppositely regulate photosystem expression
Figures
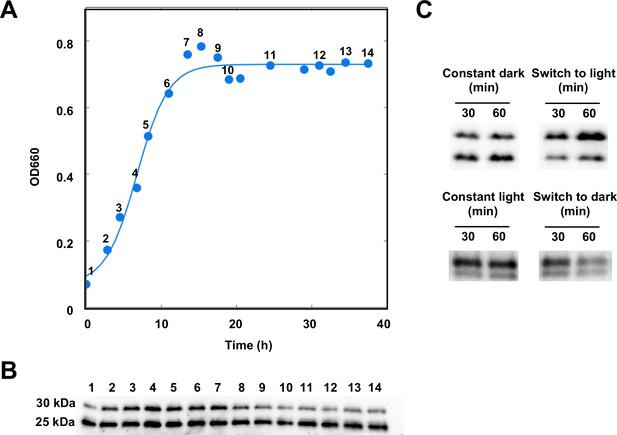
AerR exists as two forms depending on the position in a cells growth cycle.
(A) R. capsulatus grown under aerobic condition and AerR-FLAG protein expression was checked at each point (1 to 14 in growth curve). (B) Western blot analysis to detect AerR-FLAG protein. Lane numbers are responding numbers in the growth curve. (C) AerR expression was checked when cells were shifted from dark to light or from light to dark after 30 and 60 min.
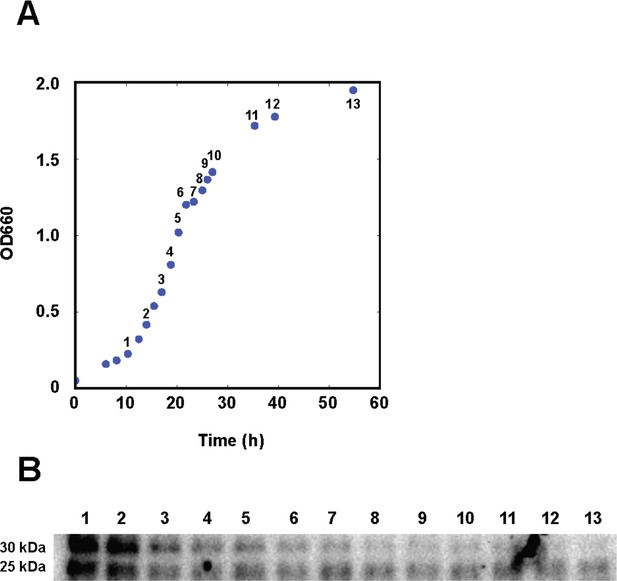
AerR also exists as two forms under photosynthetic growth conditions.
(A) R. capsulatus grown under aerobic conditions with AerR-FLAG protein expression checked at each point (1 to 13 in growth curve). (B) Western blot analysis to detect AerR-FLAG protein. Lane numbers are responding numbers in the growth curve.
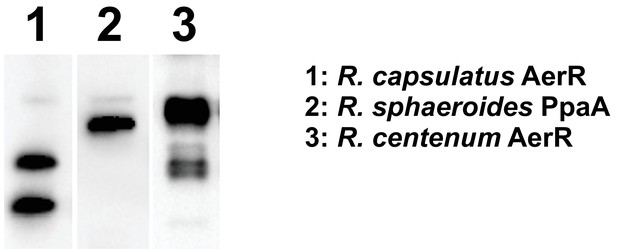
AerR (PpaA) expression pattern in other species.
Western blot analysis of AerR(PpaA)−3xFLAG protein expression in Rhodobacter sphaeroides and Rhodospirillum centenum.
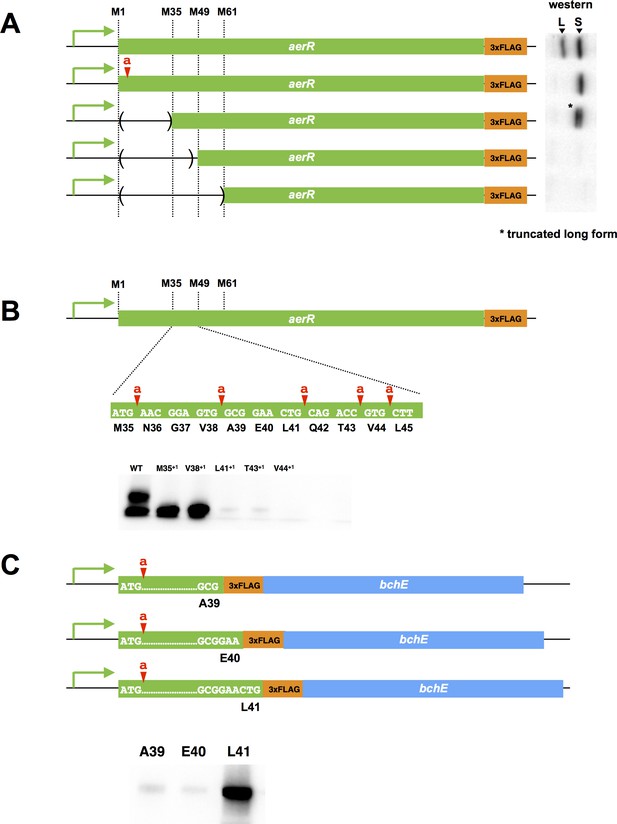
Mutational analysis of the internal second initiation codon, Leu41 (CTG).
All AerR mutant proteins were detected by Western blot analysis with anti-FLAG antibody. (A) Several truncated AerR proteins were expressed in R. capsulatus from internal start codon candidates, M35, M49, and M61, respectively. Frameshift mutations were introduced via one nucleotide insertions downstream of the Met1 codon. (B) One nucleotide was inserted after M35, V38, L41, T43, and V44 codons, respectively. (C) Three partial aerR sequences, that include the M1 codon to the A39, E40, or L41 codons, were fused to the bchE open reading frame that also had a FLAG epitope tag as a reporter. Translation from the M1 codon was blocked by a one nucleotide insertion after the M1 codon.
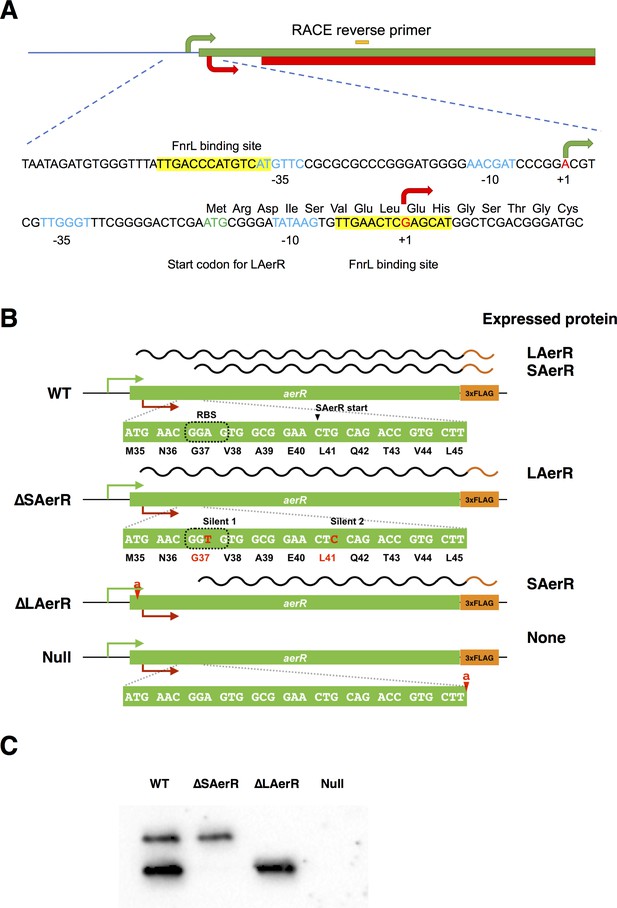
Summary of AerR translation start sites and AerR mutant construction.
(A) WT strain expresses both LAerR (green bar) and SAerR (red bar), from Met codon one and Leu codon 41 respectively. The two transcription start sites are indicted as a green arrow and a red arrow as defined by Sanger sequencing of the 5’-RACE experiments. Putative −35 and −10 promoter recognition sequences in blue with yellow highlighted sequences denoting FnrL binding sites. (B) An ∆SAerR strain that expresses only LAerR, contains two silent mutations indicated in red text. One is within an internal ribosome binding site used for short AerR translation and a second is at the SAerR initiation codon L41 codon. ∆LAerR strain that expresses only SAerR, has one nucleotide insertion downstream of M1 codon to stop LAerR translation. A null AerR strain was constructed that does not express any AerR isoform by insertion of one nucleotide downstream of the L45 codon. (C) AerR-FLAG expression pattern in the three AerR mutants along with a WT control.
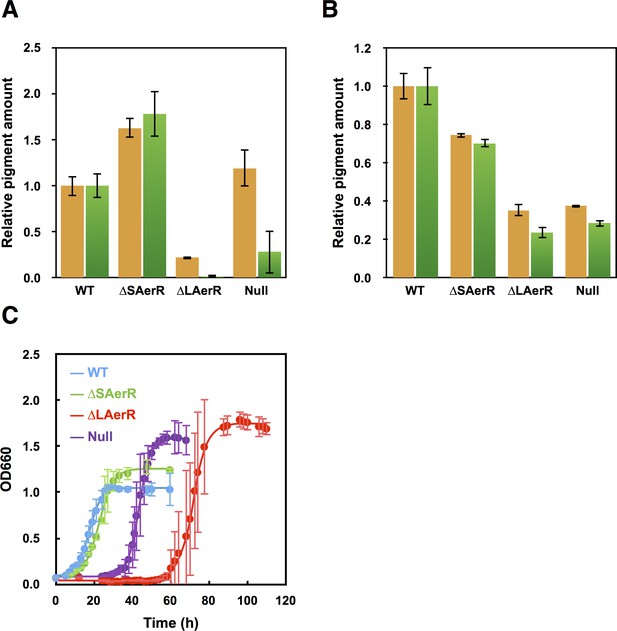
Pigment levels in WT, ∆SAerR, ∆LAerR and negative control null AerR strains and their effect on photosynthetic growth.
Total pigment obtained from organic extraction from (A) aerobically or (B) photosynthetically grown WT, ∆SAerR, ∆LAerR or negative control null AerR strains that were harvested at late-exponential phase (OD = 0.6 to 0.7). Yellow bars indicate carotenoids and green bars indicate bacteriochlorophyll. (C) Photosynthetic growth of WT (blue), ∆SAerR strain (green), ∆LAerR strain (red), and the AerR null strain (purple) after shifting from dark semi-aerobic to photosynthetic illuminated conditions.
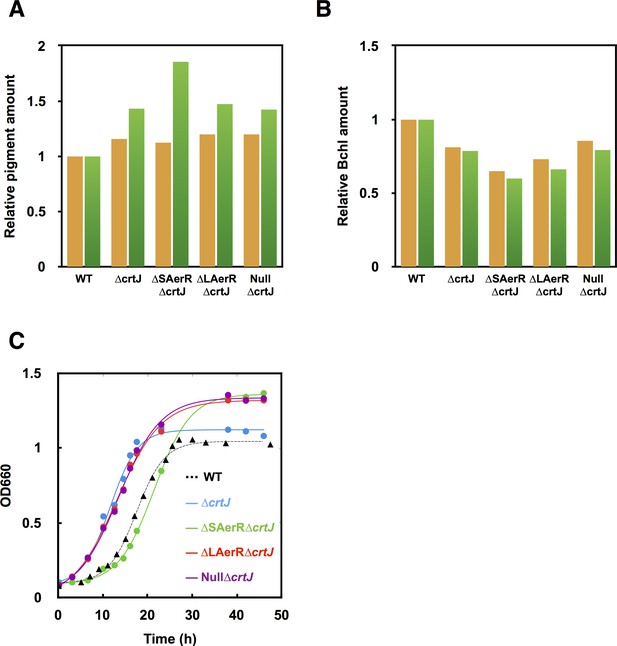
SAerR and LAerR phenotypes are suppressed by a CrtJ null mutation.
A crtJ gene deletion was constructed in ∆SAerR, ∆LAerR and ∆AerR mutant strains and then assayed for pigment production and photosynthetic growth. Bar graph of carotenoid (yellow bar) and bacteriochlorophyll (green bar) pigments present in dark semi-aerobically grown (A) or photosynthetically grown cells (B) harvested at late-exponential phase (OD = 0.6 to 0.7). (C) Growth curve of ∆crtJ (blue), ∆SAerR/∆crtJ (green), ∆LAerR/∆crtJ (red), Null/∆crtJ (purple), and WT control (black triangle) when shifted from dark aerobic to a light anaerobic conditions.
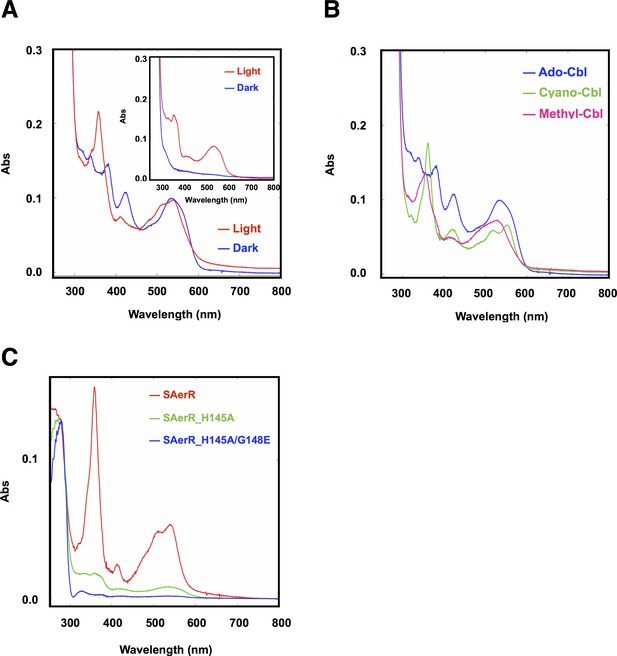
Cobalamin binding ability of SAerR protein.
(A) The insert shows a spectral scan of purified LAerR protein incubated with Ado-Cbl in dark (blue) or light (red) for 5 min followed by removal of unbound cobalamins. LAerR only binds OH-Cbl which is a product of light excitation of Ado-Cbl, thus LAerR does not bind Cbl under dark conditions. The larger spectrum is of SAerR incubated with Ado-Cbl in dark (blue) or light (red) for 5 min followed by removal of unbound cobalamins. SAerR binds both Ado-Cbl in the dark as well as light generated OH-Cbl. (B) Spectral scan of SAerR incubated with Ado-Cbl (blue), Cyano-Cbl (green), or Methyl-Cbl (pink) in dark for 5 min followed by removal of unbound cobalamins. (C) Spectral scan of purified SAerR (red), SAerR_H145A (green), and SAerR_H145A/G148E (blue) incubated with Ado-Cbl in light illumination for 5 min followed by removal of unbound cobalamins.
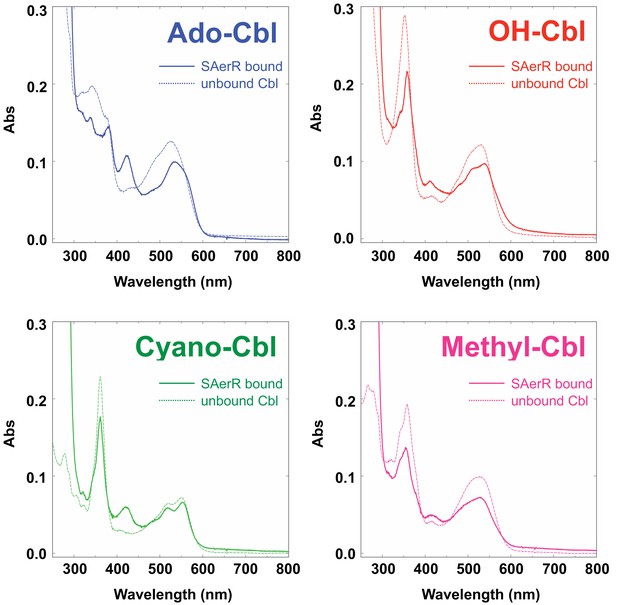
SAerR binds several kinds of cobalamin.
Spectral scan of SAerR incubated with Ado-Cbl (blue), OH-Cbl (red), Cyano-Cbl (green), or Methyl-Cbl (pink) followed by removal of unbound cobalamins. Solid lines are SAerR bound form and dotted lines are protein free of unbound cobalamins.
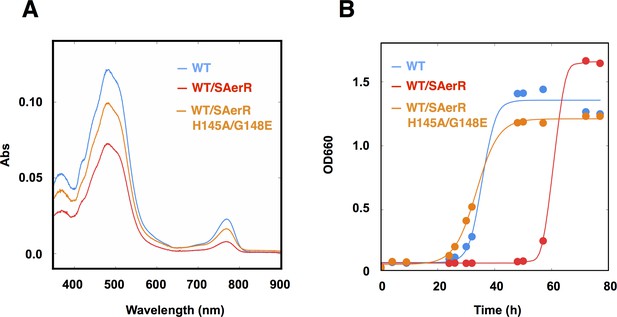
An SAerR overexpression phenotype is suppressed in vivo by mutations in the cobalamin binding motif.
(A) Spectrum of total pigment extracts from semi-aerobically grown WT strain (blue), a WT strain overexpressing SAerR (red), and a WT strain overexpressing SAerR_H145A/G148E (orange). (B) Growth curve of WT cells (blue), WT cells overexpressing SAerR (red), and WT cells overexpressing SAerR_H145A/G148E (orange). These cells were shifted from dark semi-aerobic to photosynthetic growth conditions.
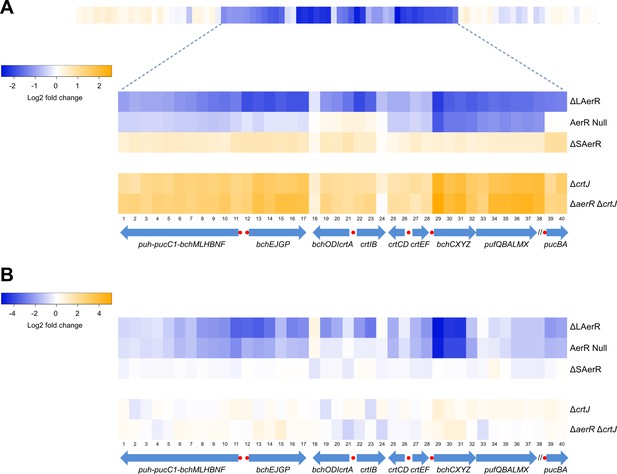
Heat map of global expression changes in AerR and CrtJ mutant strains as measured by RNA-seq.
(A) Changes in expression occuring in the photosynthesis gene cluster region of the R. capsulatus chromosome in dark semi-aerobically grown AerR mutation strains. The ΔLAerR and AerR null strains exhibit severe reduction in photosystem transcription while the ΔSAerR exhibits an increase in expression. The reduced expression phenotypes exhibited by the ΔLAerR and AerR null strains are overcome by introduction of a crtJ deletion. (B) Similar to A with the exception that the cells were grown anaerobically under photosynthetic illuminated conditions. CrtJ binding motifs are shown as a red-dots.
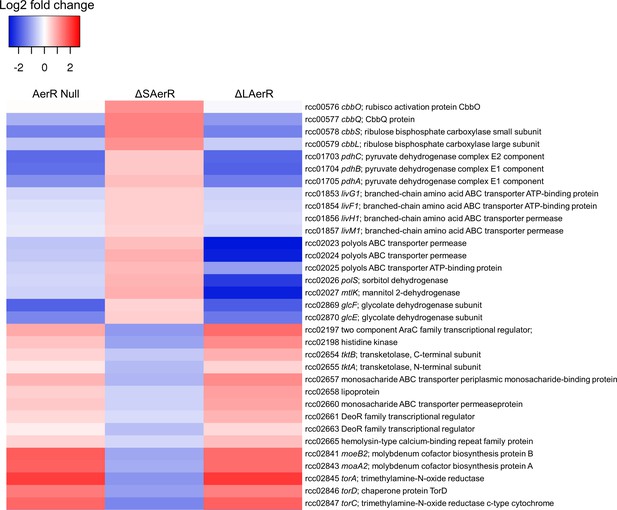
LAerR and SAerR regulates genes in opposite direction.
Although most of the genes that are affected by disruption of LAerR do not change expression level upon disruption of SAerR, there are several genes that show an opposite change when SAerR is deleted. Examples include several carbon fixation genes, several branch chain transporters genes and TMAO reductase genes.
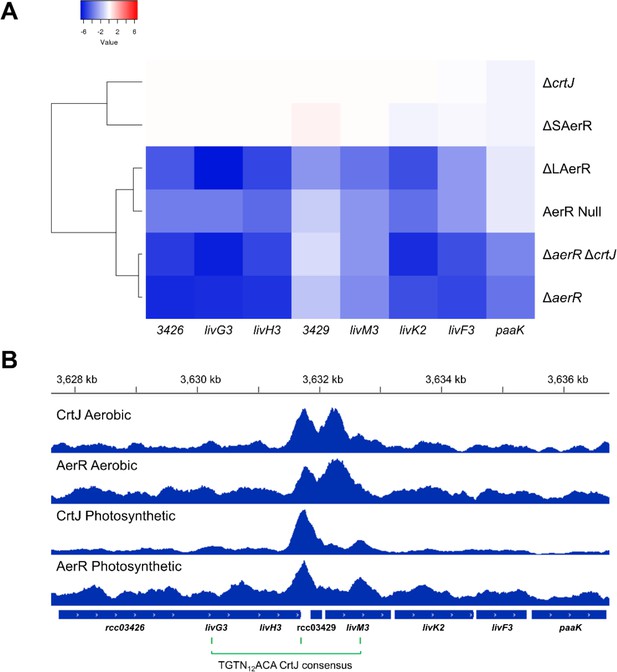
Regulation of CrtJ and AerR on the liv3 operon.
(A) ∆crtJ and ∆SAerR strains show similar liv operon expression levels (rcc03426-03433). However, ∆LaerR, aerR Null ∆aerR-∆crtJ and ∆aerR strains show highly decreased liv operon transcription under photosynthetic conditions. (B) ChIP-seq results of CrtJ and AerR in vivo binding to the liv3 operon. CrtJ and AerR co-localize to the liv promoter under both aerobic and photosynthetic conditions. Three CrtJ binding consensus sequences (TGTN12ACA) were found, with two corresponding to ChIP-seq peaks. ChIP-seq data used for this analysis was previously reported and deposited (Fang and Bauer, 2017).
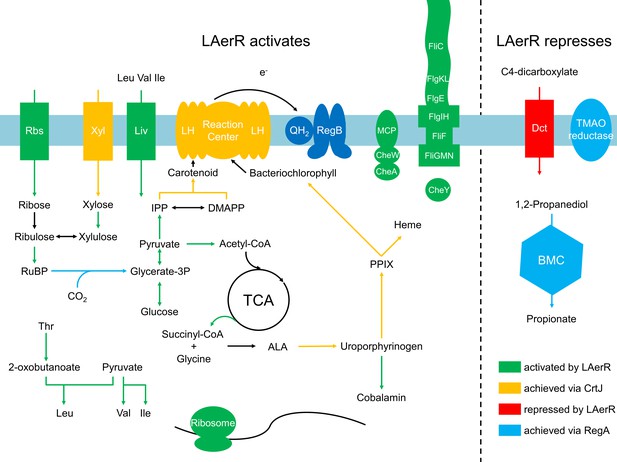
Summary of the LAerR regulon in R. capsulatus under photosynthetic growth conditions.
LAerR activates carbon fixation, chemotaxis and motility, cobalamin biosynthesis, glycolysis and TCA cycle, heme biosynthesis, photosynthesis, ribosome and some transporters like xylose, ribulose and BCAA transporters. At the same time, LAerR represses bacterial microcompartment, TMAO reductase and C4-dicarboxylate transporter. These LAerR regulatory processes involve a direct interaction of LAerR with CrtJ.
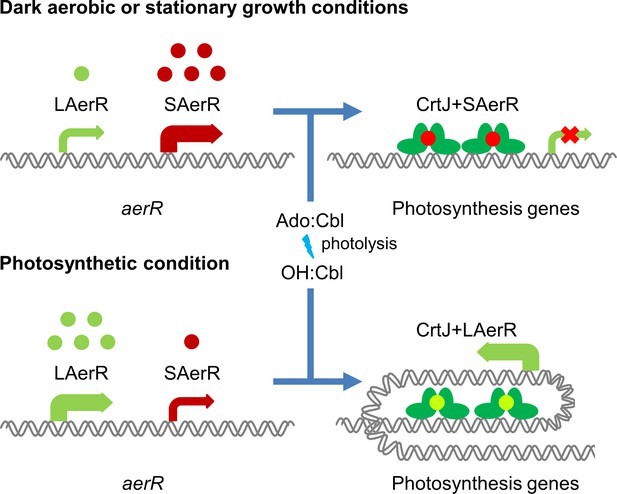
Model of LAerR and SAerR function in controlling gene expression.
The results of this study, coupled with in vivo and in vitro DNA binding studies by Fang et al (Fang and Bauer, 2017), suggest that LAerR and SAerR are both capable of interacting with CrtJ. SAerR (red dot) complexed with Adenosyl- (Ado) or Methyl-cobalamin (Me:Cbl) is predominantly made under dark and/or stationary growth conditions. SAerR complexed with these biologically active cobalamin derivatives stimulates CrtJ (dark green) mediated photosystem repression. Under photosynthetic conditions, LAerR (light green dot) is the predominate form with this variant only binding hydroxyl cobalamin (OH-Cbl) that is generated via photolysis of Ado or methyl-cobalamin. LAerR bound with OH-Cbl interacts with CrtJ in a manner that promotes extensive interaction with target promoters (Fang and Bauer, 2017) activating gene expression.

Western blot analysis of WT cell and cells that express flag tagged SAerR and flag tagged SAerR containing the Cbl binding mutations.
Antibody used is to the flag epitope.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Rhodobacter capsulatus) | WT | PMID: 1262313 | Wild type parent strain | This laboratory |
| Genetic reagent (Rhodobacter capsulatus) | ∆LAerR | This study | Strain that only expresses SAerR | |
| Genetic reagent (Rhodobacter capsulatus) | ∆SAerR | This study | Strain that only expresses LAerR | |
| Genetic reagent (Rhodobacter capsulatus) | AerR null | This study | Strain that does not express either L/SAerR | |
| Genetic reagent (Rhodobacter capsulatus) | AerR 3xFLAG | PMID: 28325764 | Strain with aerR 3xFLAG on 3' | |
| Genetic reagent (Rhodobacter capsulatus) | ∆LAerR 3xFLAG | This study | Strain that only expresses SAerR in AerR 3xFLAG background | |
| Genetic reagent (Rhodobacter capsulatus) | ∆SAerR 3xFLAG | This study | Strain that only expresses LAerR in AerR 3xFLAG background | |
| Genetic reagent (Rhodobacter capsulatus) | AerR null 3xFLAG | This study | Strain that does not express either L/SAerR in AerR 3xFLAG background | |
| Genetic reagent (Rhodobacter capsulatus) | ∆crtJ | PMID: 28325764 | Strain that does not express CrtJ | |
| Genetic reagent (Rhodobacter capsulatus) | ∆LAerR ∆crtJ | This study | Strain that only expresses SAerR in ∆crtJ background. | |
| Genetic reagent (Rhodobacter capsulatus) | ∆SAerR ∆crtJ | This study | Strain that only expresses LAerR in ∆crtJ background. | |
| Genetic reagent (Rhodobacter capsulatus) | AerR null ∆crtJ | This study | Strain that does not express either L/SAerR in ∆crtJ background. | |
| Genetic reagent (Escherichia coli) | BL21 (DE3) | NEB | C2527 | |
| Genetic reagent (Escherichia coli) | S17-1λpir | PMID: 6340113 | ||
| Genetic reagent (Escherichia coli) | HST08 | TaKaRa Bio | 9128 | |
| antibody | Anti-FLAG epitope monoclonal antibody HRP conjugate | Sigma | A8592 | |
| recombinant DNA reagent | pSUMO-CrtJ | PMID: 22715852 | ||
| recombinant DNA reagent | pSUMO-SAerR | This study | Plasmid that express SUMO-SAerR under T7 promotor. | |
| recombinant DNA reagent | pSUMO-AerR | PMID: 24329562 | Plasmid that express SUMO-LAerR under T7 promotor. | |
| recombinant DNA reagent | pBBR-aerR 3xFLAG | This study | Plasmid that express aerR-3xFLAG | |
| recombinant DNA reagent | pBBR-aerR_Met1a | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Met1 codon | |
| recombinant DNA reagent | pBBR-aerR_Met35 | This study | Plamid that express aerR-3xFLAG with a truncation from the Met1 codon to the 34th codon | |
| recombinant DNA reagent | pBBR-aerR_Met49 | This study | Plamid that express aerR-3xFLAG with a truncation from the Met1 codon to the 48th codon | |
| recombinant DNA reagent | pBBR-aerR_Met61 | This study | Plamid that express aerR-3xFLAG with a truncation from the Met1 codon to the 60th codon | |
| recombinant DNA reagent | pBBR-aerR_M35 + 1 | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Met35 codon | |
| recombinant DNA reagent | pBBR-aerR_V38 + 1 | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Val38 codon | |
| recombinant DNA reagent | pBBR-aerR_L41 + 1 | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Leu41 codon | |
| recombinant DNA reagent | pBBR-aerR_T43 + 1 | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Thr43 codon | |
| recombinant DNA reagent | pBBR-aerR_V44 + 1 | This study | Plasmid that express aerR-3xFLAG with a nucleotied A insertion after the Val44 codon | |
| recombinant DNA reagent | pBBR-aerR_A39 -FLAG-bchE | This study | Plasmid that express 3xFLAG-bchE with a partial aerR sequence from M1 to A39 as a promotor and an initiation codon | |
| recombinant DNA reagent | pBBR-aerR_E40 -FLAG-bchE | This study | Plasmid that express 3xFLAG-bchE with a partial aerR sequence from M1 to E40 as a promotor and an initiation codon | |
| recombinant DNA reagent | pBBR-aerR_L41 -FLAG-bchE | This study | Plasmid that express 3xFLAG-bchE with a partial aerR sequence from M1 to L41 as a promotor and an initiation codon | |
| peptide, recombinant protein | SUMO-CrtJ | PMID: 22715852 | ||
| peptide, recombinant protein | SUMO-LAerR | PMID: 24329562 | ||
| peptide, recombinant protein | SUMO-SAerR | This study | SUMO-tagged SAerR | |
| commercial assay or kit | In Fusion HD cloning kit | Clontech | 639648 | |
| commercial assay or kit | GeneRacer Kit | Invitrogen | 150201 | |
| commercial assay or kit | Terminator exonuclease | Epicentre | TER51020 | |
| commercial assay or kit | MST labeling kit | NanoTemper | MO-L001 | |
| software, algorithm | Trimmomatic | PMID: 24695404 | ||
| software, algorithm | Bowtie2 | PMID: 22388286 | ||
| software, algorithm | HTSeq | PMID: 25260700 | ||
| software, algorithm | DESeq2 | PMID: 20979621 |
Additional files
-
Supplementary file 1
This file shows expression for all genes in the R. capsulatus genome listed in numerical order for the AerR null strain, the strain lacking SAerR and the strain lacking LAerR.
The expression changes were determined relative to expression levels observed in the parent wild type strain using RNA-seq. Cells were grown under dark semi-aerobic conditions. p-adjusted value <0.01 are shown in red.
- https://doi.org/10.7554/eLife.39028.018
-
Supplementary file 2
This file shows expression for all genes in the R. capsulatus genome listed in numerical order for the AerR null strain, the strain lacking SAerR and the strain lacking LAerR.
The expression changes were determined relative to expression levels observed in the parent wild type strain using RNA-seq. Cells were grown under illuminated anaerobic photosynthetic conditions. p-adjusted value <0.01 are shown in red.
- https://doi.org/10.7554/eLife.39028.019
-
Supplementary file 3
This file contains a list of all genes in the AerR null strain that undergo significant changes in expression as based on a p-adjusted value <0.01.
The gene expression changes are shown as log 2-fold changes and were measured using RNA-seq. The first tab shows all genes undergoing changes in expression in cells grown dark semi-aerobically while the second tab shows changes in expression in cells grown under illuminated anaerobic photosynthetic conditions.
- https://doi.org/10.7554/eLife.39028.020
-
Supplementary file 4
This file contains a list of all genes that undergo significant changes in expression as based on a p-adjusted value <0.01.
The gene expression changes are shown as log 2-fold changes and were measured using RNA-seq. The strains assayed are the AerR null strain, a strain lacking SAerR (DSAerR) and a strain lacking LAerR (DLAerR). All strains were grown under illuminated anaerobic photosynthetic conditions. Note that genes are clustered into groups of similar function with individual tabs.
- https://doi.org/10.7554/eLife.39028.021
-
Supplementary file 5
This file lists the sequence of all PCR primers used in this study for plasmid and mutant construction.
- https://doi.org/10.7554/eLife.39028.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39028.023





