Stepwise wiring of the Drosophila olfactory map requires specific Plexin B levels
Figures
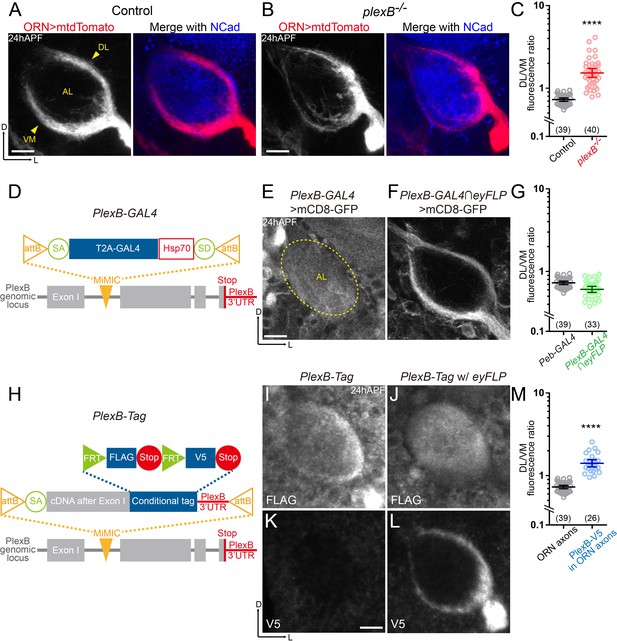
ORN axons of the DL trajectory express a higher level of PlexB proteins than the VM axons.
(A) In wild-type pupal brains at 24 hr after puparium formation (24hAPF), ORN axons bifurcate and form the dorsolateral (DL) and ventromedial (VM) trajectories (arrowheads) circumscribing the antennal lobe (AL). ORN axons were labeled by the pan-ORN Peb-GAL4 (Sweeney et al., 2007) driven mtdTomato expression. Antennal lobes were co-stained with a neuropil marker N-cadherin (NCad). (B) In plexB homozygous mutant, ORN axons preferentially choose the DL trajectory. (C) Fluorescence intensity ratios of ORN axon trajectories (DL/VM) in wild-type and plexB–/– brains at 24hAPF. Geometric means: control, 0.73; plexB–/–, 1.53. (D) Design of the PlexB-GAL4: a T2A-GAL4 cassette (Diao et al., 2015) was inserted into a MiMIC locus in the first coding intron of PlexB. SA, splicing acceptor. Hsp70, terminator sequence of Hsp70. SD, splicing donor (not functional because mRNAs terminate at the Hsp70 terminator). (E) PlexB-GAL4 labels the antennal lobe and nearby brain structures at 24hAPF. The signal from ORNs is not detectable. Dotted circle, antennal lobe (AL). (F) Intersection of PlexB-GAL4 and eyFLP (FLP in ORNs) labels both DL and VM trajectories. (G) DL/VM fluorescence intensity ratios of the pan-ORN GAL4 (Peb-GAL4) and the PlexB-GAL4 intersected with eyFLP. Geometric means: Peb-GAL4, 0.73; PlexB-GAL4∩eyFLP, 0.61. (H) Design of the PlexB conditional tag: a cassette including a splicing acceptor (SA), the PlexB coding sequence after the first exon, FRT-FLAG-Stop-FRT-V5-Stop, and the PlexB 3’-UTR sequence, was inserted into a MiMIC locus in the first coding intron of PlexB. (I, J) FLAG staining of 24hAPF brains of PlexB-Tag alone (I) or PlexB-Tag with eyFLP (J). (K, L) V5 staining of 24hAPF brains of PlexB-Tag alone (K) or PlexB-Tag with eyFLP (L). (M) DL/VM fluorescence intensity ratios: ORN axon distributions were calculated based on the Peb-GAL4 > mtdTomato signal (as in Figure 1A); ORN-specific PlexB protein distributions were calculated based on the V5 staining of PlexB-Tag with eyFLP (as in Figure 1L). Geometric means: ORN axon distribution, 0.73; ORN-specific PlexB protein distribution, 1.41. Sample sizes are noted in parentheses. Significance between two groups was determined by an unpaired, two-tailed t-test. ****p<0.0001. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
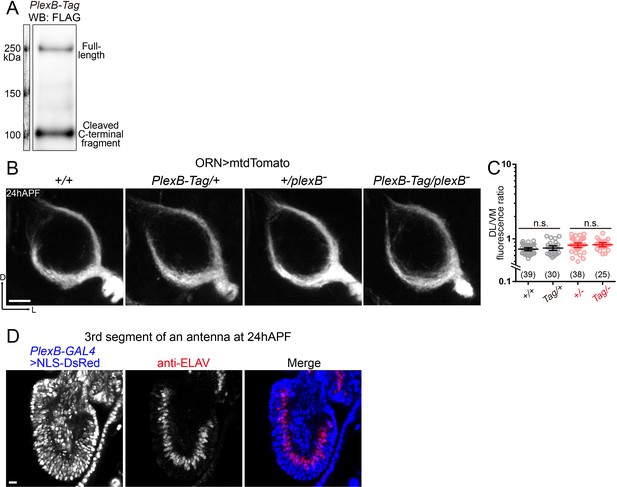
PlexB-Tag functions normally as the wild-type PlexB, and PlexB-GAL4 labels vast majority of ORNs at 24hAPF.
(A) Western blotting of FLAG-tagged PlexB proteins in 24hAPF brains shows the stable expression of PlexB, as well as the expected processing by cleavage. No abnormal degradation was observed. (B, C) PlexB-Tag functions normally as the wild-type PlexB (marked as ‘+') in ORN axon trajectory formation at 24hAPF. (B) Representative images of each genotype. (C) Fluorescence intensity ratios of ORN axon trajectories (DL/VM). Geometric means: +/+, 0.73; Tag/+, 0.75; +/–, 0.82; Tag/–, 0.84. (D) PlexB-GAL4 labels ORNs (ELAV+) and other cells (ELAV–) in the third segment of an antenna at 24hAPF. Vast majority of ORNs (92.3%; 287 out of 311 counted) express PlexB. NLS-DsRed: DsRed fused to a nuclear localization sequence. Sample sizes are noted in parentheses. Significance between two groups was determined by an unpaired, two-tailed t-test. n.s., not significant. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
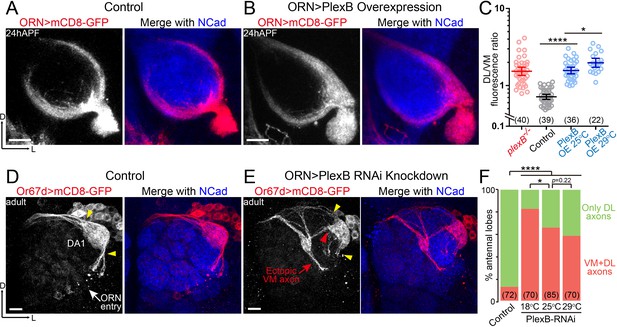
ORN axon trajectory choice requires specific levels of PlexB.
(A) ORN axon trajectories in wild-type flies at 24hAPF. (B) Overexpression of PlexB in ORNs shifts ORN axons to the DL trajectory. (C) Fluorescence intensity ratios of ORN axon trajectories (DL/VM). Geometric means: plexB-/-, 1.53; control, 0.71; PlexB OE at 25°C, 1.57; PlexB OE at 29°C, 1.98. (D) ORN axons targeting to the DA1 glomerulus were labeled by membrane-tethered GFP driven by Or67d-QF. In wild-type flies, DA1 ORN axons stay only at the DL edge of the antennal lobe (yellow arrowheads). Or67d-QF has non-specific labeling of several non-neuronal cells outside of each antennal lobe. No axon or dendrite was observed from these cells. (E) PlexB RNA interference (RNAi) in ORNs leads to the formation of a VM axon bundle of DA1 ORNs (red arrow). A subset of ectopic VM axons innervates the DA1 glomerulus at a medial entry point (red arrowhead), instead of the normal entry points at the edge of the antennal lobe (yellow arrowheads). (F) Quantification of antennal lobes with the VM axon bundle of DA1 ORNs in wild-type flies and PlexB RNAi flies at different temperatures. Sample sizes are noted in parentheses. Significance among multiple groups in Figure 2C was determined by one-way ANOVA with Tukey’s test for multiple comparisons. Significance of the contingency table in Figure 2F was determined by Fisher's exact test. *p<0.05; ****p<0.0001. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
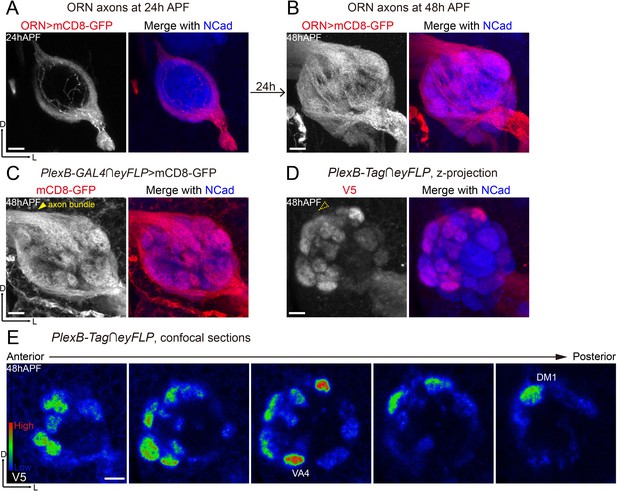
Differential distribution of PlexB proteins in ORN axons at the stage of glomerular selection.
(A) At 24hAPF, ORN axons bifurcate and form the DL and VM trajectories circumscribing the antennal lobe. ORN axons were labeled by the pan-ORN Peb-GAL4 driver line (Sweeney et al., 2007). (B) By 48hAPF, ORN axons have innervated the antennal lobe, selected targeting areas, and formed proto-glomeruli with prospective PN partners. Within 24 hr (24hAPF to 48hAPF), the antennal lobe is significantly enlarged and ORN axons have extensively elaborated terminal arbors. (C) Intersection of PlexB-GAL4 and eyFLP (FLP in ORNs) labels nearly all glomeruli. The generic membrane-anchored mCD8-GFP uniformly labels axons, including the commissure bundle (arrowhead). (D, E) At 48hAPF, ORN axons targeting to several discrete glomeruli express higher levels of PlexB proteins than their neighbors. These PlexB-high glomeruli include VA4, DM1, and a dorsal one whose identity cannot be unambiguously determined at this stage. (D) Maximum z-projection of a confocal stack. Glomerular boundaries are visible while the commissure axon bundle is barely observed (dotted arrowhead). (E) Single confocal sections from anterior to posterior. Colors represent the intensity of V5 signal: red, high; green, intermediate; blue, low. Images are shown as maximum z-projections of confocal stacks, except for Figure 3E, where single confocal sections are shown. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
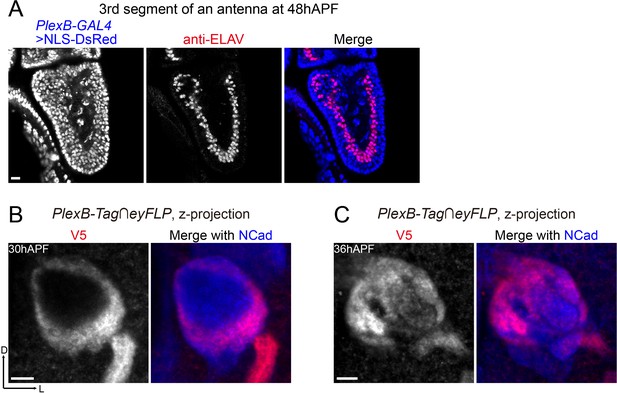
PlexB-GAL4 labels almost all ORNs at 48hAPF, and PlexB proteins in ORN axons undergo dynamic changes between 30hAPF and 36hAPF.
(A) PlexB-GAL4 labels ORNs (ELAV+) and other cells (ELAV–) in the third segment of an antenna at 48hAPF. Most ORNs (98.7%; 312 out of 316 counted) express PlexB. NLS-DsRed: DsRed fused to a nuclear localization sequence. (B, C) PlexB proteins in ORN axons at 30hAPF and 36hAPF, respectively. At 30hAPF, PlexB is still concentrated in ORN axons as they begin to invade the interior of the antennal lobe. At 36hAPF when ORN axons innervate the antennal lobe, PlexB is already preferentially concentrated in the medial antennal lobe, similar to the distribution at 48hAPF.
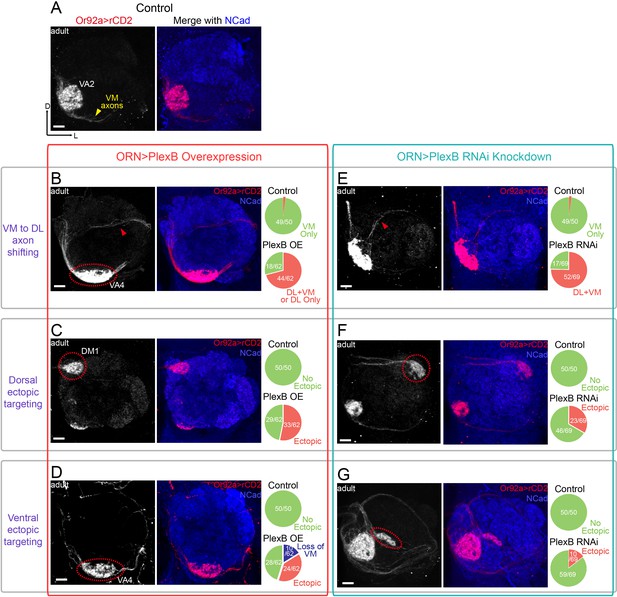
ORN glomerular selection requires specific levels of PlexB.
(A) ORN axons targeting to the VA2 glomerulus were labeled by membrane-localized rCD2 driven by an Or92a promoter. Or92a+ ORN axons predominantly choose the VM trajectory (yellow arrowhead). (B) Some Or92a+ ORN axons choose the DL trajectory (red arrowhead) in the PlexB overexpression flies. In the shown antennal lobe, Or92a+ ORN axons targeting to the VA4 glomerulus are also observed (dotted red circle). (C) PlexB overexpression causes Or92a+ ORN axons to ectopically target to dorsal glomeruli (dotted red circle), mainly DM1 (67% of dorsal mis-targeting cases). (D) PlexB overexpression causes Or92a+ ORN axons to ectopically innervate ventral glomeruli (dotted red circle), mainly VA4 (88% of ventral mis-targeting cases). (E) Some Or92a+ ORN axons choose the DL trajectory (red arrowhead) when PlexB is knocked down by RNAi in ORNs. (F) PlexB knockdown causes Or92a+ ORN axons to incorrectly target to dorsolateral glomeruli (dotted red circle), distinct from the stereotyped DM1 mis-targeting in the PlexB overexpression flies. (G) In a few PlexB knockdown flies, local ectopic targeting near VA2 was observed (dotted red circle). However, mis-targeting to VA4, a predominant phenotype in PlexB overexpression, was not observed. Antennal lobe counts of each phenotype and sample sizes are noted in the pie charts. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
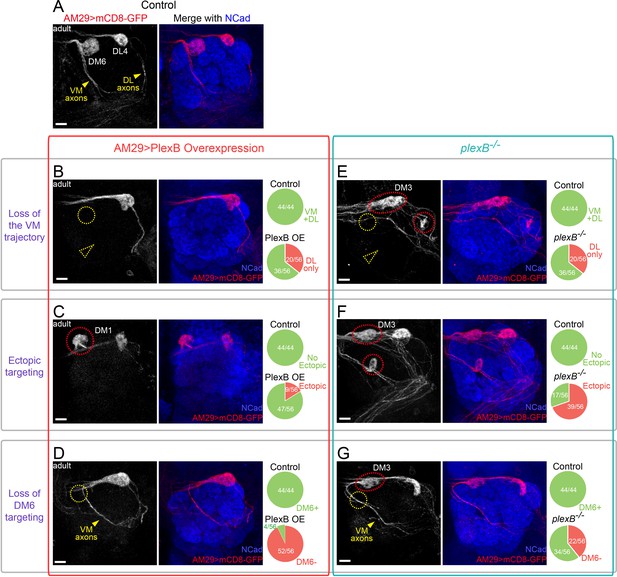
Glomerular selection of AM29+ ORNs requires specific levels of PlexB.
(A) ORN axons innervating the DL4 and DM6 glomeruli were labeled by membrane-tethered GFP driven by AM29-GAL4. DL4 and DM6 ORN axons choose the DL and VM trajectories, respectively (yellow arrowheads). (B) Overexpression of PlexB by AM29-GAL4 causes the loss of the VM trajectory (dotted yellow arrowhead) and the loss of DM6 glomerular targeting (dotted yellow circle). (C) PlexB overexpression causes ectopic targeting to dorsal glomeruli (dotted red circle), mainly DM1 (56% of mis-targeting cases). (D) In more than half of the antennal lobes in the PlexB overexpression flies, DM6 glomerular targeting disappears (dotted yellow circle) although VM axons still exist and pass by the vicinity of the putative DM6 area (yellow arrowhead). (E) VM axons disappear (dotted yellow arrowhead) in about 1/3 of plexB mutant brains. Loss of DM6 glomerular targeting (dotted yellow circle), as well as ectopic glomerular targeting (dotted red circles), was also observed in this example antennal lobe. (F) In plexB mutants, AM29+ ORN axons ectopically target to DM3 and other glomeruli (dotted red circles; DM3: 51% of mis-targeting cases) but not DM1. (G) plexB mutants lose DM6 glomerular innervation (dotted yellow circle), even when VM axons are present and pass by the vicinity of DM6 (yellow arrowhead). Ectopic targeting to DM3 is also observed in the shown antennal lobe (dotted red circle). Antennal lobe counts of each phenotype and sample sizes are noted in the pie charts. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
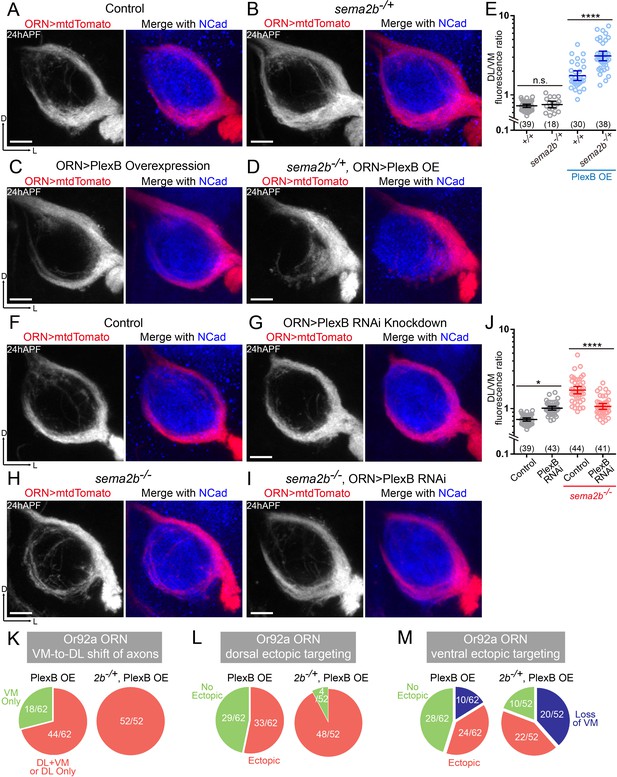
Sema2b antagonizes the effect of high-level PlexB.
(A) ORN axon trajectories of wild-type flies at 24hAPF. (B) sema2b heterozygous flies have normal ORN axon trajectories. (C) PlexB overexpression shifts ORN axons to the DL trajectory. (D) sema2b heterozygosity (sema2b–/+) enhances the DL shift of ORN axons caused by PlexB overexpression. (E) Fluorescence intensity ratios of ORN axon trajectories (DL/VM). Geometric means: +/+, 0.73; sema2b–/+, 0.76; PlexB OE in +/+, 1.75; PlexB OE in sema2b–/+, 3.10. (F) ORN axon trajectories of wild-type flies at 24hAPF. (G) PlexB knockdown by RNA interference (RNAi) in ORNs slightly shifts ORN axons to the DL trajectory, phenotypically resembling plexB mutants but with less severity. (H) In sema2b mutant flies, ORN axons preferentially choose the DL trajectory at 24hAPF. (I) PlexB RNAi in ORNs suppresses the DL shift of ORN axons caused by the loss of Sema2b. (J) Fluorescence intensity ratios of ORN axon trajectories (DL/VM). Geometric means: control, 0.73; PlexB RNAi, 1.01; sema2b–/–, 1.83; PlexB RNAi in sema2b–/–, 1.11. (K, L, M) sema2b heterozygosity (sema2b–/+) enhances both incorrect axon trajectory choice and ectopic glomerular targeting caused by PlexB overexpression. Each phenotype is described in detail in Figure 4. (K) VM-to-DL shift of ORN axons, as in Figure 4B; (L) dorsal ectopic targeting, as in Figure 4C; (M) ventral ectopic targeting, as in Figure 4D. Sample sizes are noted in parentheses. Antennal lobe counts of each phenotype and sample sizes are noted in the pie charts. Significance among multiple groups was determined by one-way ANOVA with Tukey’s test for multiple comparisons. n.s., not significant; *p<0.05; ****p<0.0001. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
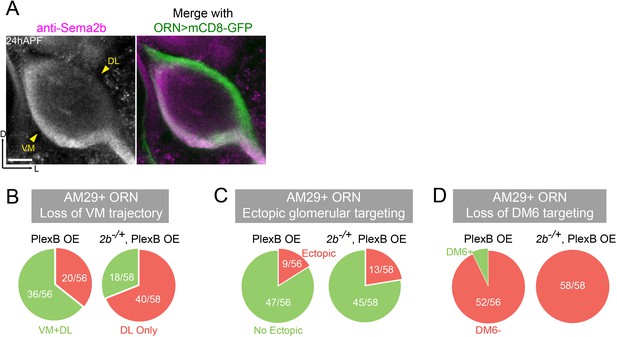
(A) At 24hAPF, Sema2b proteins are enriched in the VM ORN axon bundle. (B, C, D) sema2b heterozygosity (sema2b–/+) enhances both incorrect axon trajectory choice and ectopic glomerular targeting caused by PlexB overexpression in AM29+ ORNs. Each phenotype is described in detail in Figure 4—figure supplement 1. (B) loss of the VM trajectory, as in Figure 4—figure supplement 1B; (C) ectopic glomerular targeting, as in Figure 4—figure supplement 1C; (D) loss of the glomerular targeting to DM6, as in Figure 4—figure supplement 1D. Antennal lobe counts of each phenotype and sample sizes are noted in the pie charts. Images are shown as maximum z-projections of confocal stacks. Scale bars, 10 μm. Axes, D (dorsal), L (lateral).
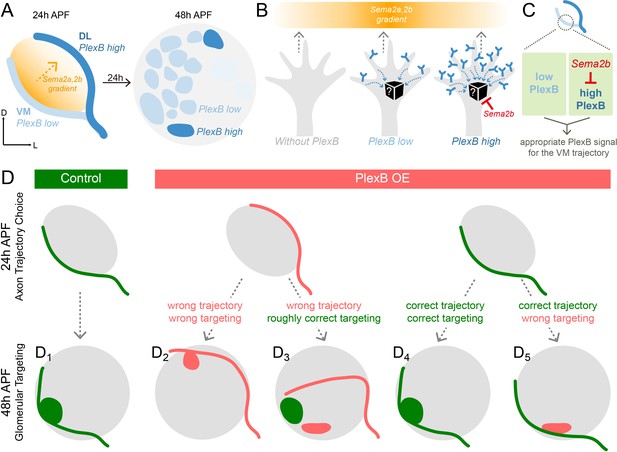
Schematic summary of the roles of PlexB in the stepwise assembly of the Drosophila olfactory circuit.
(A) At 24hAPF, ORN axons of the DL trajectory express a higher level of PlexB proteins, compared to the VM axons. Sema2a and Sema2b proteins form a descending gradient from VM to DL in the antennal lobe (Joo et al., 2013). Within the next 24 hr, ORN axons enter the antennal lobe and form proto-glomeruli with PN dendrites. At 48hAPF, ORN axons innervating several discrete glomeruli express PlexB proteins at high levels, exhibiting a different distribution pattern from that of 24hAPF. (B) ORN axons without PlexB (as in the plexB mutant) or with high-level PlexB (as the wild-type DL axons or the VM axons in PlexB overexpression) preferentially choose the low-Sema2a/2b area (DL). Only ORN axons with low-level PlexB (as the wild-type VM axons or the DL axons in PlexB knockdown) target to the high-Sema2a/2b area (VM). Although it is unknown how PlexB levels affect its signaling (black box), Sema2b antagonizes the activity of high-level PlexB. (C) ORN axons destined to the VM trajectory express low levels of PlexB. Additionally, Sema2b is enriched in the VM bundle to antagonize the signal of high-level PlexB. These two mechanisms thus ensure an appropriate PlexB signal for the formation of the VM trajectory. (D) ORN glomerular selection is affected by, but also can be independent of, the preceding trajectory choice. Although ectopic glomerular targeting frequently occurs when the trajectory goes wrong (D2), a portion of ORN axons can overcome the initial trajectory choice and return to the roughly correct area (D3). Among ORN axons achieving correct trajectory choice, many innervate nearby incorrect glomeruli if the PlexB level is disturbed (D5).
Tables
| Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|
| Pebbled-GAL4 | doi:10.1016/j.neuron.2006.12.022 | ||
| AM29-GAL4 | doi:10.1038/nn1832 | ||
| Or67d-QF | doi:10.1016/j.neuron.2013.06.014 | ||
| Or92a-rCD2 | this study | Or92a promoter fused to a rat CD2 coding sequence in a P-element vector | |
| eyFLP3.5 | doi:10.1016/j.neuron.2005.09.019 | ||
| UAS-mtdTomato | doi:10.1016/j.cell.2010.02.025 | RRID:BDSC_30124 | |
| UAS-mCD8-GFP | doi:10.1016/S0896-6273 (00)80701–1 | RRID:BDSC_5130 | |
| UAS-FRT-Stop-FRT-mCD8-GFP | doi:10.1038/nn.2442 | RRID:BDSC_30125 | |
| QUAS-mCD8-GFP | doi:10.1016/j.cell.2010.02.025 | RRID:BDSC_30002 | |
| UAS-RedStinger | doi:10.2144/04363ST03 | RRID:BDSC_8547 | |
| plexBKG00878 | doi:10.1534/genetics.104.026427 | RRID:BDSC_14579 | |
| PlexBMI15559 | doi:10.1038/nmeth.1662 | RRID:BDSC_61730 | |
| PlexB-Tag | this study | Endogenous conditional tagging of PlexB | |
| PlexB-GAL4 | Xiaojun Xie and Alex Kolodkin, unpublished | Coding intron MiMIC-T2A-GAL4 of PlexB | |
| UAS-PlexB-RNAi | doi:10.1038/nmeth.1592 | RRID:BDSC_57813 | |
| UAS-PlexB | doi:10.1016/j.neuron.2013.03.022 | ||
| sema2bf02042 | doi:10.1038/ng1314 | RRID:BDSC_18505 | |
| UAS-Sema2b | doi:10.1016/j.neuron.2011.02.050 | ||
| rat anti-Ncad | Developmental Studies Hybridoma Bank | RRID:AB_528121 | 1:40 in 5% normal donkey serum |
| mouse anti-ELAV | Developmental Studies Hybridoma Bank | RRID:AB_528217 | 1:40 in 5% normal donkey serum |
| chicken anti-GFP | Aves Labs | RRID:AB_10000240 | 1:1000 in 5% normal donkey serum |
| rabbit anti-DsRed | Clontech | RRID:AB_10013483 | 1:200 in 5% normal donkey serum |
| mouse anti-rat CD2 | Bio-Rad | RRID:AB_321238 | 1:200 in 5% normal donkey serum |
| mouse anit-FLAG | Sigma-Aldrich | RRID:AB_439685 | 1:100 in 5% normal donkey serum |
| mouse anti-V5 | Thermo Fisher | RRID:AB_2556564 | 1:100 in 5% normal donkey serum |
| rabbit anti-Sema2b | doi:10.1016/j.neuron.2011.09.026 | 1:500 in 5% normal donkey serum | |
| ZEN | Carl Zeiss | RRID:SCR_013672 | |
| ImageJ | National Institutes of Health | RRID:SCR_003070 | |
| Prism | GraphPad | RRID:SCR_002798 | |
| Photoshop | Adobe | RRID:SCR_014199 | |
| Illustrator | Adobe | RRID:SCR_010279 |
Additional files
-
Supplementary file 1
Genotypes of flies in each experiment.
- https://doi.org/10.7554/eLife.39088.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39088.013





