Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase
Figures
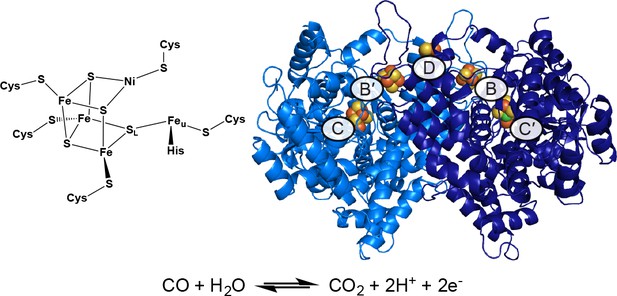
Structure of the C-cluster and CODH.
Protein structure shown in ribbon representation in blue with metalloclusters labeled and shown as spheres; Ni in green, Fe in orange, S in yellow. Reaction catalyzed by CODH is shown below.
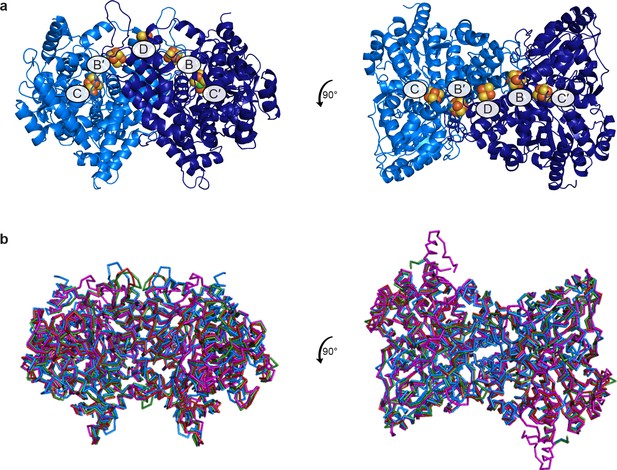
Overall structure of DvCODH.
(a) Structure of DvCODH in ribbon representation with metalloclusters shown as spheres; Ni in green, Fe in orange, S in yellow. (b) Overlay of the Cα traces of DvCODH (blue), Rhodospirillum rubrum CODH (red; PDB ID: 1JQK), Carboxydothermus hydrogenoformans CODH (green; PDB ID: 1SU8), and Moorella thermoacetica CODH (purple; PDB ID: 1MJG).
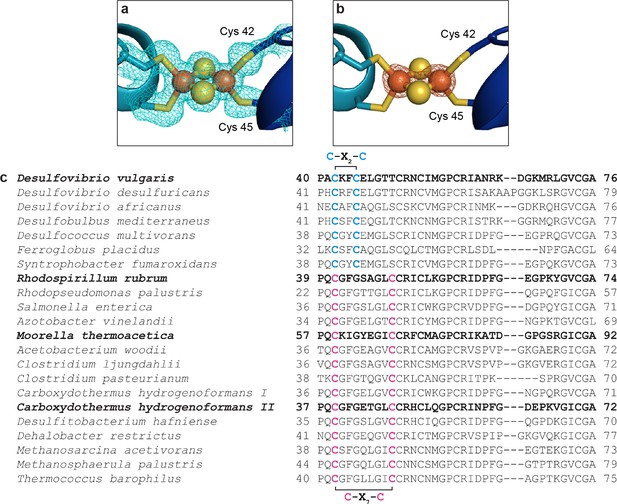
DvCODH contains a distinguishing [2Fe-2S] D-cluster.
(a) Electron density (2FO−FC, contoured to 1σ) for the [2Fe-2S] cluster of DvCODH. Protein is shown in ribbon representation with ligating amino acid residues as sticks and [2Fe-2S] cluster as spheres and sticks; Fe in orange, S in yellow. (b) Fe anomalous difference map (contoured to 8σ) at the D-cluster supports the assignment of a [2Fe-2S] cluster in this position. (c) Alignment of CODH sequences. The C-X2-C D-cluster binding motif observed in the DvCODH sequence is also present in the sequences of other, uncharacterized CODHs. Sequences of structurally and biochemically characterized CODHs are in bold.
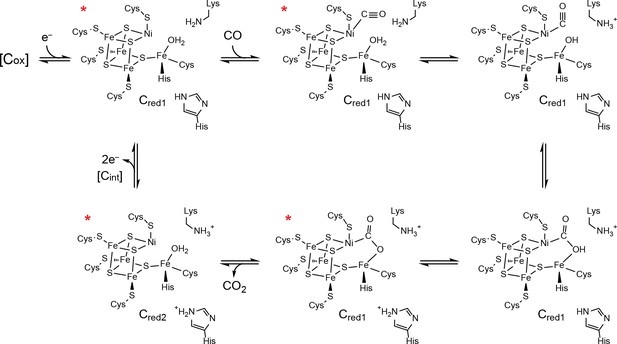
Proposed mechanism of CO oxidation at the CODH C-cluster.
The catalytic cycle begins with the C-cluster in the Cred1 redox state (a one-electron reduced state of the C-cluster) with H2O bound to Feu (Jeoung and Dobbek, 2007; Kung et al., 2009). CO binds to the cluster in a bent binding mode (Gong et al., 2008; Kung et al., 2009) and then undergoes a ‘carbon shift,’ positioning the carbonyl carbon atom for nucleophilic attack by a Feu-bound hydroxide, formed by loss of a proton from Feu-bound H2O to a catalytic base, which is proposed to be a conserved lysine residue (Drennan et al., 2001; Dobbek et al., 2001; Kim et al., 2004). The resulting COOH-type species is deprotonated by a second catalytic base, proposed to be an active site histidine residue, to form a metallocarboxylate species (Drennan et al., 2001; Dobbek et al., 2001; Jeoung and Dobbek, 2009; Fesseler et al., 2015; Chen et al., 2003; Kim et al., 2004). CO oxidation reduces the C-cluster by two electrons forming the Cred2 state (a species that is two-electrons more reduced than Cred1) and CO2 is released (Jeoung and Dobbek, 2007; Kumar et al., 1993; Anderson and Lindahl, 1996). We note that, although Cred2 formation and CO2 release have been drawn concomitantly, the rate of CO2 release has been shown to be slower than the rate of cluster reduction (Seravalli and Ragsdale, 2008). For the next round of turnover, the cluster undergoes a two-electron oxidation, reforming the Cred1 state. Cluster oxidation is thought to proceed through two single-electron transfer events via an intermediate Cint redox state as electrons flow through the B- and D-clusters to an external redox partner, such as ferredoxin. The Cox redox state is one electron more oxidized than Cred1. States of the C-cluster that have been visualized crystallographically are indicated with a red asterisk.
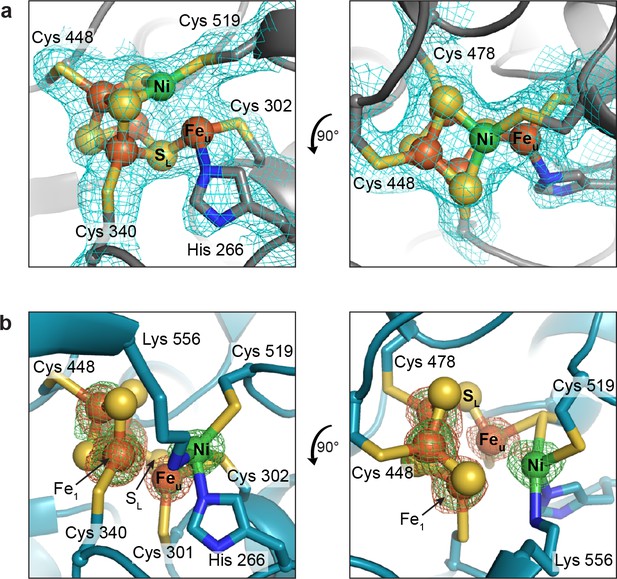
The DvCODH C-cluster adopts alternative conformations.
(a) C-cluster with simulated annealing composite omit electron density map (contoured to 1σ) as observed in the 2.50 Å resolution structure of as-isolated DvCODH (batch 1). The cluster adopts the canonical conformation, ligated by the indicated amino acid residues. Lys556 does not ligate to the cluster in this conformation and has been omitted here for clarity. The positioning of Lys556 is shown in Figure 3—figure supplement 1a. (b) C-cluster as observed in the 1.72 Å resolution structure of as-isolated DvCODH (batch 2). Ni, Feu, and SL have shifted relative to the canonical cluster and Cys301 and Lys556 form new interactions to the cluster. Fe and Ni anomalous difference maps, calculated from data collected at Fe and Ni peak wavelengths (7130 and 8360 eV, respectively), are shown as green and orange mesh contoured to 6σ and 5σ, respectively. Protein is shown in ribbon representation with ligating amino acid residues as sticks and C-cluster as spheres and sticks; Ni in green, Fe in orange, S in yellow, N in blue.
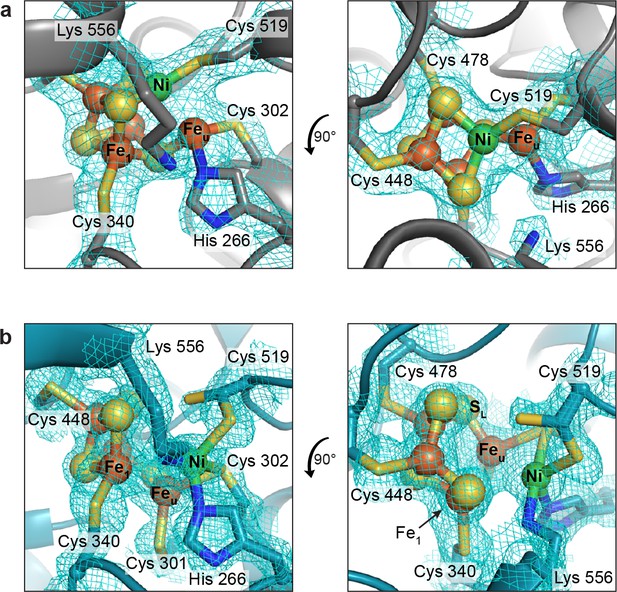
Views of the alternative C-cluster arrangements.
(a) The canonical C-cluster as observed in protein batch 1. Lys556 has been shown for comparison with its position in the non-canonical cluster. (b) The C-cluster as observed in protein batch 2. Clusters are depicted as shown in Figure 3 with simulated annealing composite omit electron density maps contoured to 1σ.
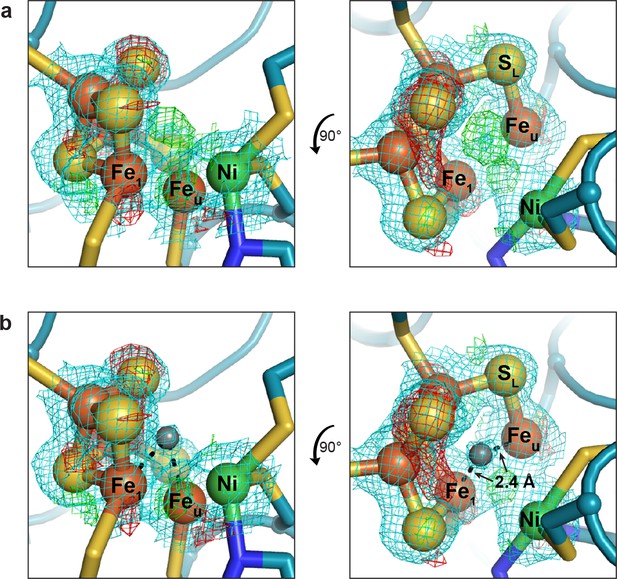
A peak of residual electron density is present within the alternative C-cluster.
(a) View of the cluster prior to modeling and refinement of a ligand into the residual electron density. The peak of positive difference electron density (green mesh) indicates the presence of a ligand at a site bridging Feu and Fe1. (b) View of the cluster following refinement of an unknown ligand (grey sphere) into the electron density. The unknown ligand is within 2.4 Å of both Feu and Fe1. Protein is shown as in Figure 3. The lysine residue has been omitted from the left hand panels for simplicity. 2FO−FC electron density (blue mesh) contoured to 1σ; FO−FC electron density (green and red mesh) contoured to ±3σ.
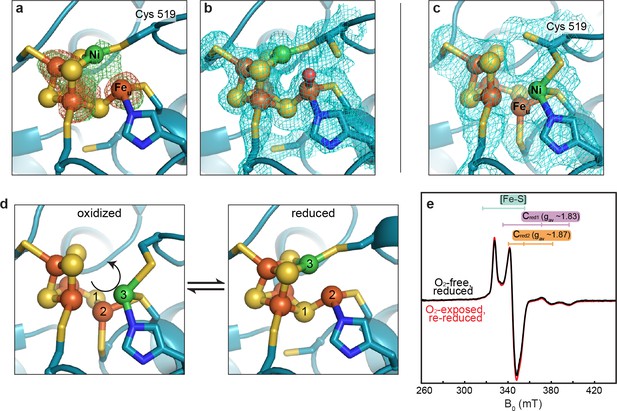
The DvCODH C-cluster undergoes reversible, redox-dependent conformational changes.
(a) C-cluster of dithionite-soaked DvCODH crystal (batch 2). Ni and Fe anomalous difference maps are shown as green and orange mesh, respectively, and contoured to 5σ. (b) C-cluster of dithionite-soaked DvCODH crystal (batch 2) with simulated annealing composite omit electron density map contoured to 1σ. A water molecule (red sphere) is observed bound to Feu. (c) C-cluster of reduced and then air-exposed DvCODH crystal (batch 2) with simulated annealing composite omit electron density map contoured to 1σ. Lys556 has not been shown for simplicity. (d) Illustration of the conversion between the oxidized and reduced states of the C-cluster. Arrow and numbers indicate the assumed direction of metal ion movement from the oxidized state to the reduced state. (e) Continuous-wave X-Band EPR spectra of dithionite reduced DvCODH before (black trace) and after (red trace) exposure to air. Experimental conditions: mw power = 0.2 mW, mw frequency = 9.34 GHz, modulation amplitude 1 mT, temperature = 10 K. For panels a-d, protein is shown in ribbon representation with ligating amino acid residues as sticks and C-cluster as spheres and sticks; Ni in green, Fe in orange, S in yellow, N in blue, O in red.
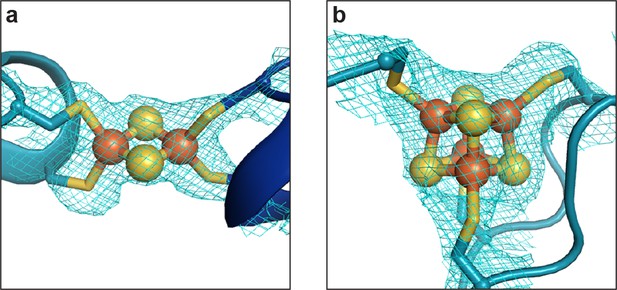
The D- and B-clusters of DvCODH are resistant to oxidative damage.
(a) Electron density at the D-cluster following 2 d at ambient atmospheric conditions. (b) Electron density at the B-cluster following 2 d at ambient atmospheric conditions. Protein is shown in ribbon representation with ligating amino acid residues as sticks and Fe-S clusters as spheres and sticks; Fe in orange, S in yellow. Simulated annealing composite omit electron density maps contoured to 1σ.
The C-cluster of DvCODH undergoes redox-dependent conformational changes.
In this movie, the C-cluster begins in the oxidized state, morphs into the canonical, reduced state, and then returns to the oxidized state. Cluster is shown in ball and stick representation; Ni in green, Fe in orange, S in yellow. Protein is shown in ribbon representation with ligating amino acid residues in sticks; S in yellow, N in blue.
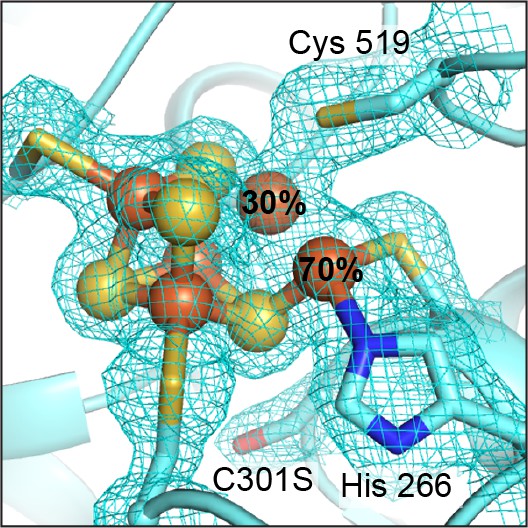
The DvCODH(C301S) C-cluster is a mixture of species.
The C-cluster has been refined with an alternative conformation of Feu. At 70% occupancy, Feu is ligated by His266 and Cys302 in its canonical binding site. At 30% occupancy, Feu is incorporated into the Fe-S cubane portion of the cluster. 2FO−FC electron density contoured to 1σ. Protein is shown in ribbon representation with ligating amino acid residues as sticks and C-cluster as spheres and sticks; Fe in orange, S in yellow, N in blue, O in red.
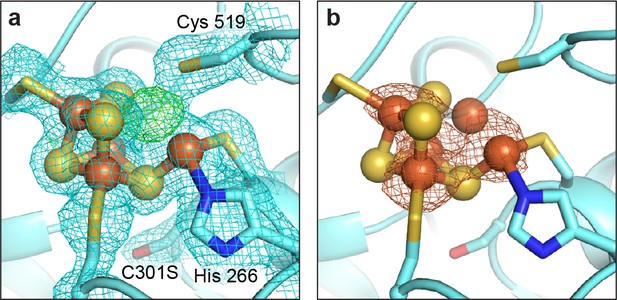
The DvCODH(C301S) C-cluster is a mixture of species.
(a) Refinement of a [3Fe-4S]-Feu C-cluster results in positive FO−FC electron density (green mesh, contoured to +3σ) at the Ni binding site. 2FO−FC electron density (blue mesh) contoured to 1σ. (b) Fe anomalous difference map (orange mesh, contoured to 4.5σ) indicates the presence of Fe at partial occupancy in the canonical Ni binding site. Protein is shown as in Figure 5.
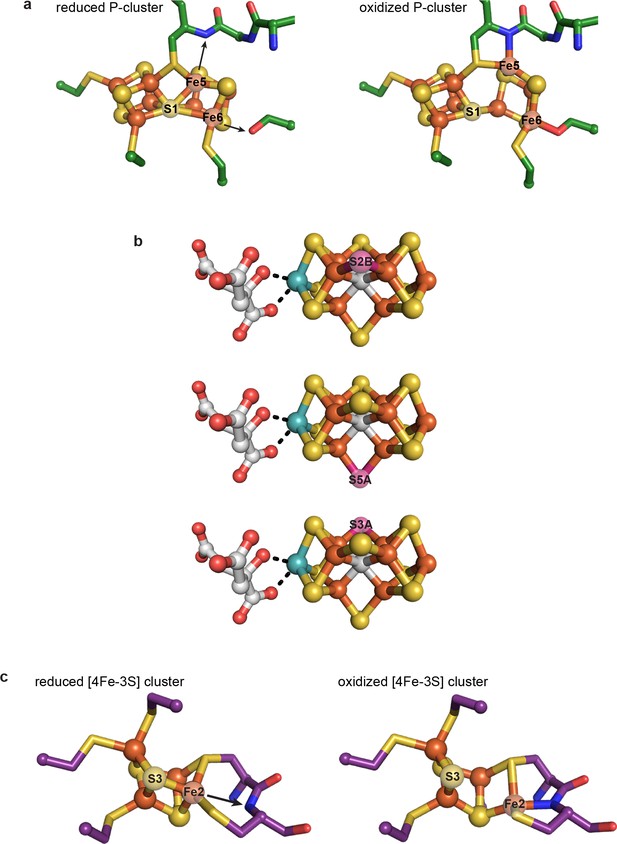
Previously characterized structural lability in Fe-S clusters.
(a) The reduced and oxidized P-cluster of nitrogenase. Upon oxidation, Fe5 and Fe6 lose coordination to the central S1 ion, moving distances of 1.4 and 0.9 Å, respectively. Arrows in the left hand panel show the direction of Fe ion movement. Protein is shown as sticks with P-cluster as spheres and sticks; Fe in orange, S in yellow, C in green, N in blue, O in red. PDB IDs: 3MIN (reduced) and 2MIN (oxidized). (b) The FeMo-cofactor of nitrogenase undergoes turnover-dependent rearrangements, observed as the movement of an artificially-incorporated Se atom. Under turnover conditions, Se migrates through the cluster from position S2B to S5A to S3A. FeMo-cofactor is shown as spheres and sticks; Fe in orange, S in yellow, Se in pink, Mo in teal, C in grey, O in red. PDB ID: 5BVG. (c) A unique [4Fe-3S] cluster is present in the O2-tolerant membrane-bound hydrogenase. Upon oxidation, Fe2 loses coordination to S3 and becomes coordinated by a backbone amide group of the protein. Arrow in the left hand panel shows the direction of Fe ion movement. Protein is shown as sticks with [4Fe-3S] cluster as spheres and sticks; Fe in orange, S in yellow, C in purple, N in blue, O in red. PDB IDs: 3AYX (reduced), 2AYY (oxidized).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Desulfovibrio vulgaris str. Hildenborough) | cooS | NA | NCBI:2795474 | |
| Gene (Desulfovibrio vulgaris str. Hildenborough) | cooC | NA | NCBI:2795475 | |
| Cell line (Desulfovibrio fructosovorans str. MR400) | Desulfovibrio fructosovorans str. MR400 | PMID:1943706 | ||
| Recombinant DNA reagent | modified pBGF4 shuttle vector | PMID:26255854 | ||
| Sequence- based reagent | C301S forward primer | Eurogentec | ACATCAACGTGGCGGGG CTATCCTGCACGGGTA ACGAACTGCTC | |
| Sequence- based reagent | C301S reverse primer | Eurogentec | GAGCAGTTCGTTACCCG TGCAGGATAGCCCCGCC ACGTTGATGT | |
| Peptide, recombinant protein | DvCODH | PMID:26255854 | ||
| Peptide, recombinant protein | DvCODH(C301S) | this paper | DvCODH variant produced in the lab of Dr. C. Léger as described in Methods | |
| Software, algorithm | XDS/XSCALE | PMID:20124692 | RRID:SCR_015652 | |
| Software, algorithm | Phaser | PMID:19461840 | RRID:SCR_014219 | |
| Software, algorithm | Scupltor | PMID:21460448 | ||
| Software, algorithm | Schwarzenbacher algorithm | PMID:15213384 | ||
| Software, algorithm | Coot | PMID:20383002 | RRID:SCR_014222 | |
| Software, algorithm | Phenix | PMID:20124702 | RRID:SCR_014224 | |
| Software, algorithm | TLS parameterization | PMID:16552146 | ||
| Software, algorithm | MolProbity | PMID:20057044 | RRID:SCR_014226 | |
| Software, algorithm | PyMOL | www.pymol.org/ | RRID:SCR_000305 | |
| Software, algorithm | EasySpin | PMID:16188474 |
Additional files
-
Supplementary file 1
Crystallographic data collection and refinement statistics.
- https://doi.org/10.7554/eLife.39451.016
-
Supplementary file 2
Metal content and activity of DvCODH preparations.
- https://doi.org/10.7554/eLife.39451.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39451.018





