Efficient single-copy HDR by 5’ modified long dsDNA donors
Figures
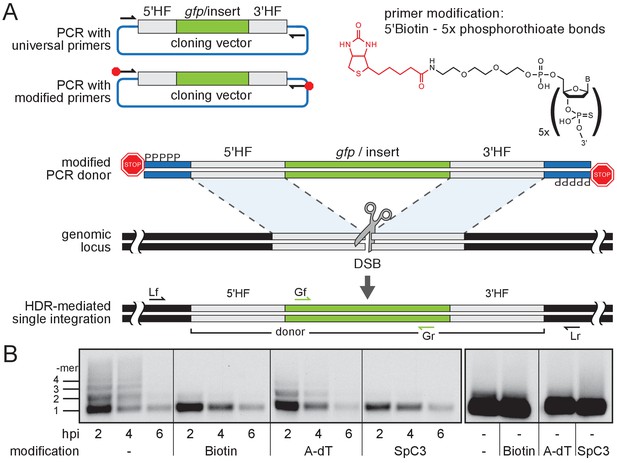
Modification of 5’ ends of long dsDNA fragments prevents in vivo multimerization.
(A) Schematic representation of long dsDNA donor cassette PCR amplification with universal primers (black arrows) complementary to the cloning vector backbone outside of the assembled donor cassette (e. g. gfp with homology flanks). Bulky moieties like Biotin at the 5’ ends of both modified primers (red octagon) prevent multimerization/NHEJ of dsDNA, providing optimal conditions for HDR-mediated single-copy integration following CRISPR/Cas9-introduced DSB at the target locus (grey scissors). Representation of locus (Lf/Lr) and internal gfp (Gf/Gr) primers for PCR genotyping of putative HDR-mediated gfp integration events. (B) Southern blot analysis reveals the monomeric state of injected dsDNA fragments in vivo for 5’ modification with Biotin or Spacer C3. Long dsDNAs generated with control unmodified primers or Amino-dT attached primers multimerize as indicated by a high molecular weight ladder apparent already within two hours post-injection (hpi). Note: 5’ moieties did not enhance the stability of injected DNA.
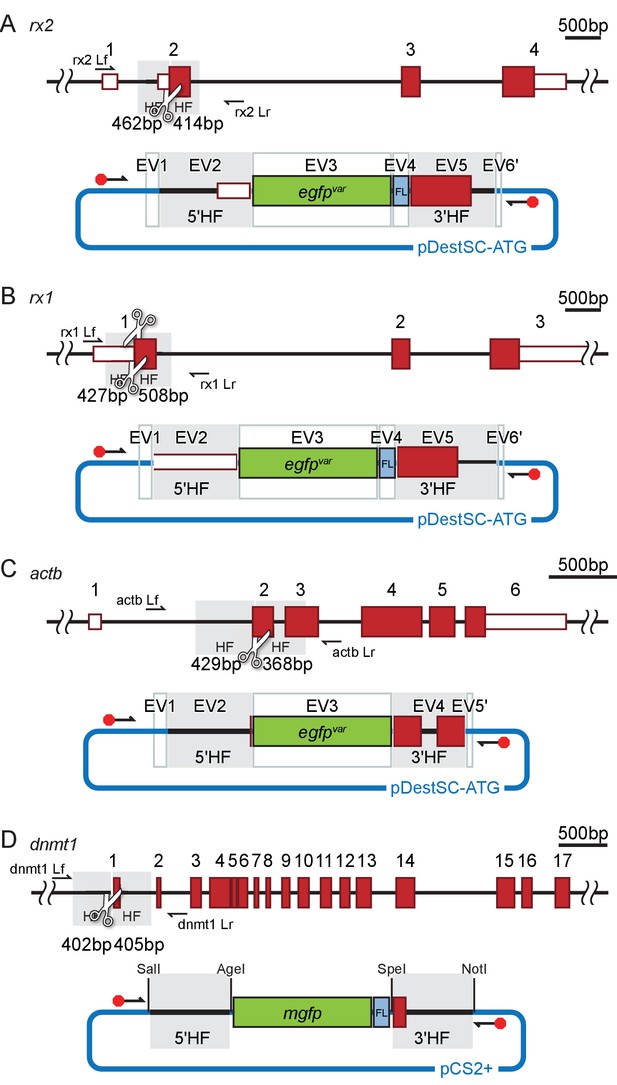
Schematic representation of the donor plasmids.
(A–D) Schematic to-scale representation of respective target locus (A, rx2; B, rx1; C, actb; D, dnmt1) with UTR (white boxes with red outlines) and exons (red boxes) highlighted. Homology flanks (HF, grey), sgRNA target sites (white scissors) and respective locus primers (black arrows, Lf, Lr, see Supplementary file 2) are indicated. Respective assembled donor plasmid (A, B, C Golden GATEway (Kirchmaier et al., 2013) or D, conventional cloning) that served as a template for PCR amplification of the unmodified/modified long dsDNA gfp donor cassette is depicted below (FL, flexible linker; backbone sequence in blue). Entry vectors (EV) or restriction enzyme sites for cloning are indicated. The position of primers flanking the donor cassette is indicated, modification highlighted by a red octagon.
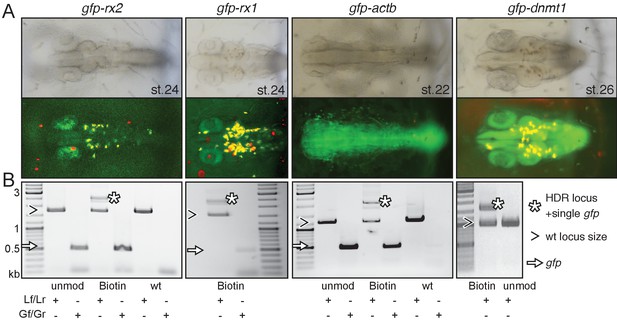
Modification of 5’ ends of long dsDNA fragments promotes HDR-mediated single-copy integration.
(A) GFP expression in the respective expression domain after HDR-mediated integration of modified dsDNA gfp donor cassettes into rx2, rx1, actb and dnmt1 ORFs in the injected generation. (B) Individual embryo PCR genotyping highlights efficient HDR-mediated single-copy integration of 5’Biotin modified long dsDNA donors, but not unmodified donor cassettes. Locus PCR reveals band size indicative of single-copy gfp integration (asterisk) besides alleles without gfp integration (open arrowhead). Amplification of gfp donor (white arrow) for control.
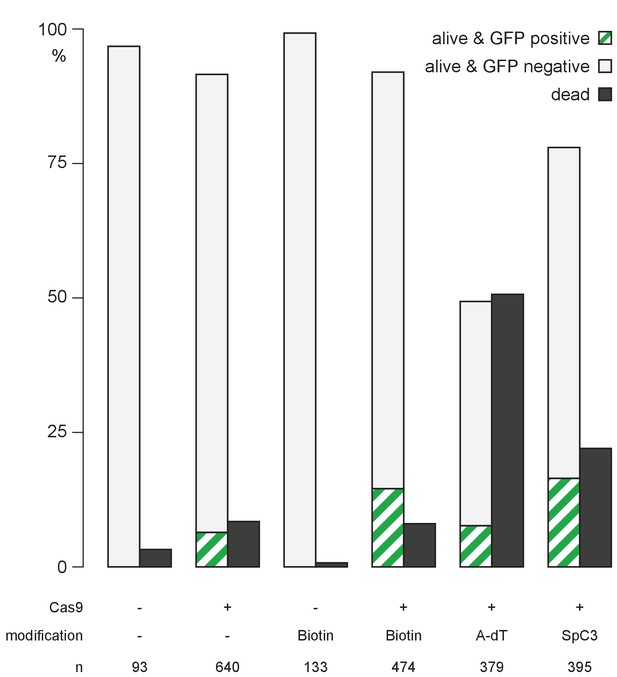
Quantification of survival and GFP expression of injected embryos.
Embryos injected with unmodified or 5’Biotin, Amino-dT or Spacer C3 modified long dsDNA gfp-rx2 donor cassettes were scored for survival at one dpi, and for GFP expression at two dpi. n, total number of injected embryos.
-
Figure 2—figure supplement 1—source data 1
Quantification of survival and GFP expression of injected embryos.
- https://doi.org/10.7554/eLife.39468.007
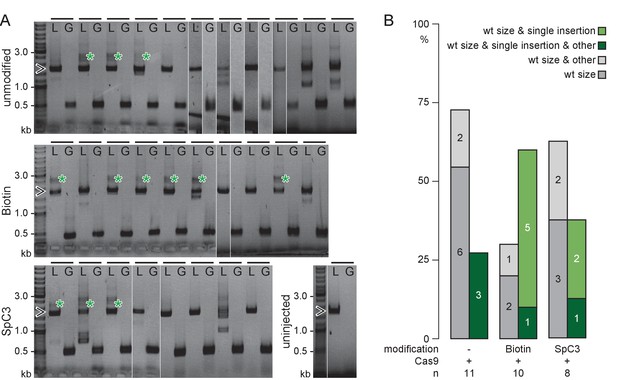
5’Biotin modification of long dsDNA donors strongly enhances HDR-mediated integration.
(A) rx2 locus PCR genotyping of individual GFP-Rx2 positive embryos injected with unmodified or 5’Biotin or Spacer C3 modified long dsDNA gfp donor cassettes (green asterisk, single-copy HDR-mediated integration of gfp, 2547 bp, open arrowhead, rx2 allele without gfp integration, 1719 bp;). Horizontal bar, individual embryo; L, rx2 locus PCR with rx2 Lf/rx2 Lr; G, gfp internal PCR for control with Gf/Gr. (B) Qualitative summary of band spectrum (single-copy HDR-mediated gfp integration, rx2 allele without gfp integration, other) resulting from PCR genotyping in (A). n, number of genotyped GFP-Rx2 expressing embryos.
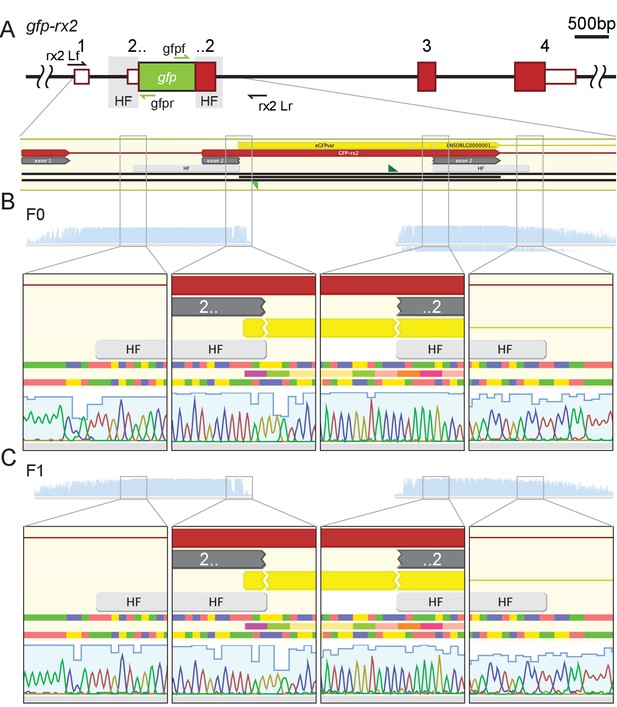
Stable germline transmission of the single-copy HDR-mediated precise gfp integration.
(A) Schematic to-scale representation of the gfp-rx2 locus with UTR (white boxes with red outlines), exons (red boxes) and homology flanks (HF, grey) highlighted. (B, C) Individual GFP-Rx2 expressing embryos (F0, B; F1, C) were genotyped using primers rx2 Lf/rx2 Lr (black arrows) and sequenced with gfpf and gfpr primers (green arrows).
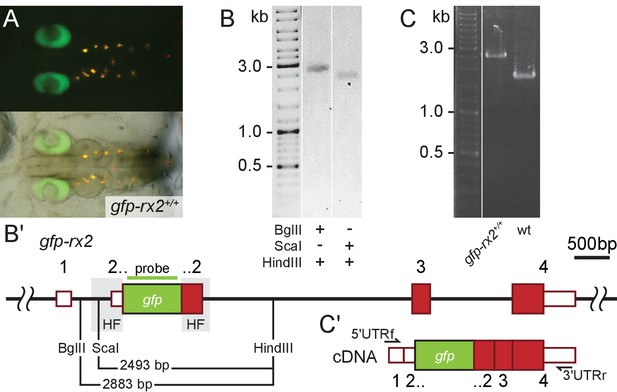
Single-copy integration of long dsDNA donor establishes stably transmitted gfp-rx2 fusion gene.
(A) F2 homozygous embryos exhibit GFP-Rx2 fusion protein expression in the pattern of the endogenous gene in the retina. (B) Southern Blot analysis of F2 gfp-rx2 embryos reveals a single band for a digestion scheme cutting outside the donor cassette (BglII/HindIII) or within the 5’ donor cassette and in intron 2 (ScaI/HindIII) indicating precise single-copy donor integration. (B’) Schematic representation of the modified locus indicating the restriction sites and the domain complementary to the probe used in (B). (C) RT-PCR analysis on mRNA isolated from individual homozygous F3 embryos indicates the transcription of a single gfp-rx2 fusion mRNA in comparison to the shorter wild-type rx2 mRNA as schematically represented in (C’).
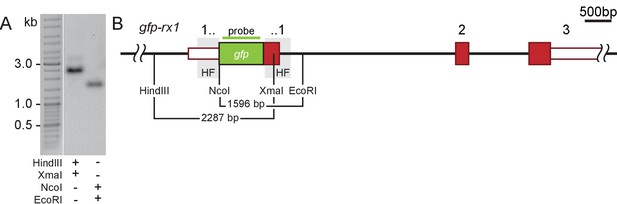
Stably transmitted single-copy integration of the gfp-rx1 donor cassette.
(A) Southern Blot analysis of F2 gfp-rx1 embryos reveals a single band for a digestion scheme cutting outside the donor cassette and within the 3’ donor cassette (HindIII/XmaI) or within the 5’ donor cassette and in intron 1 (NcoI/EcoRI) indicating precise single-copy donor integration. (A’) Schematic representation of the modified locus indicating the restriction sites and the domain complementary to the probe used in (A).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Oryzias latipes) | Cab | other | medaka Southern wild-type population | |
| Strain, strain background (Oryzias latipes) | rx2-gfp | this paper | ||
| Strain, strain background (Oryzias latipes) | rx1-gfp | this paper | ||
| Strain, strain background (Oryzias latipes) | actb-gfp | this paper | ||
| Strain, strain background (Oryzias latipes) | dnmt1-gfp | this paper | ||
| Recombinant DNA reagent | rx2-gfp donor cassette | this paper | ||
| Recombinant DNA reagent | rx1-gfp donor cassette | this paper | ||
| Recombinant DNA reagent | actb-gfp donor cassette | this paper | ||
| Recombinant DNA reagent | dnmt1-gfp donor cassette | this paper | ||
| Sequence- based reagent | rx2 5'HF f | this paper | with BamHI restriction site: GCCGGATCCAAGCATGTCAAAACGTAGAAGCG | |
| Sequence- based reagent | rx2 5'HF r | this paper | with KpnI restriction site: GCCGGTACCCATTTGGCTGTGGACTTGCC | |
| Sequence- based reagent | rx2 3'HF f | this paper | with BamHI restriction site: GCCGGATCCCATTTGTCAATGGAC ACGCTTGGGATGGTGGACGAT | |
| Sequence- based reagent | rx2 3'HF r | this paper | with KnpI restriction site: GCCGGTACCTGGACTGGACTGGAAGTTATTT | |
| Sequence- based reagent | rx2 sgRNA f | this paper | substituted nucleotides to facilitate T7 in vitro transcription of the sgRNA oligonucleotides are shown in small letters TAgGCATTTGTCAATGGATACCC | |
| Sequence- based reagent | rx2 sgRNA r | this paper | AAACGGGTATCCATTGACAAATG | |
| Sequence- based reagent | rx2 Lf/5’UTRf | this paper | TGCATGTTCTGGTTGCAACG | |
| Sequence- based reagent | rx2 Lr | this paper | AGGGACCATACCTGACCCTC | |
| Sequence- based reagent | actb 5’HF f | this paper | with BamHI restriction site: GGGGATCCCAGCAACGACTTCGCACAAA | |
| Sequence- based reagent | actb 5’HF r | this paper | with KnpI restriction site: GGGGTACCGGCAATGTCATCATCCATGGC | |
| Sequence- based reagent | actb 3’HF f | this paper | with BamHI restriction site: GGGGATCCGACGACGATATAGCTG CACTGGTTGTTGACAACGGATCTG | |
| Sequence- based reagent | actb 3’HF r | this paper | with KnpI restriction site: GGGGTACCCAGGGGCAATTCTCAGCTCA | |
| Sequence- based reagent | actb sgRNA f | this paper | TAGGATGATGACATTGCCGCAC | |
| Sequence- based reagent | actb sgRNA r | this paper | AAACGTGCGGCAATGTCATCAT | |
| Sequence- based reagent | actb Lf | this paper | GTCCGAGTTGAGGGTGTCTG | |
| Sequence- based reagent | actb Lr | this paper | CATGTGCTCCACTGTGAGGT | |
| Sequence- based reagent | dnmt1 5’HF f | this paper | with SalI restriction site: AATTTGTCGACGCTTTGA CAGTTAACCTACACG | |
| Sequence- based reagent | dnmt1 5’HF r | this paper | with AgeI restriction site: AATTTACCGGTCGTAACTGCA AACTAAAAAATAAAAC | |
| Sequence -based reagent | dnmt1 3’HF f | this paper | with SpeI restriction site: AATTTACTAGTATGCCATCCAGAA CGTCCTTATCTCTACCAGACGATG TCAGAAAAAGGTAC | |
| Sequence- based reagent | dnmt1 3’HF r | this paper | with NotI restriction site: AATTTGCGGCCGCCTACACATA TTGTCTGTGATAC | |
| Sequence- based reagent | mgfpf | this paper | with AgeI restriction site: AATTTACCGGTACTAGTACCATG AGTAAAGGAGAAGAACTTTTCAC | |
| Sequence- based reagent | mgfpr | this paper | with SpeI restriction site: AATTTACTAGTCGCGGCTGCACTT CCACCGCCTCCCGATCCGCCACC GCCAGAGCCACCTCCGCCTGAAC CGCCTCCACCGCTCAGGCTAGCTT TGTATAGTTCATCCATGCCATG | |
| Sequence- based reagent | dnmt1 sgRNA f | this paper | substituted nucleotides to facilitate T7 in vitro transcription of the sgRNA oligonucleotides are shown in small letters TAgGACATCGTCTGGCAAAGAC | |
| Sequence- based reagent | dnmt1 sgRNA r | this paper | AAACGTCTTTGCCAGACGATGT | |
| Sequence- based reagent | dnmt1 Lf | this paper | CTCAATGTAAACACTTCGTGTCGCTTC | |
| Sequence -based reagent | dnmt1 Lr | this paper | TTGCATGCATATTCAAAGTTGTCAAAG | |
| Sequence- based reagent | rx1 5’HF f | this paper | with BamHI restriction site: GCCGGATCCGCATCCGAAAGG TAAGGACTGCAAACC | |
| Sequence- based reagent | rx1 5’HF r | this paper | with KpnI restriction site: GCCGGTACCCATGAGAGCG TCTGGGCTCTGACC | |
| Sequence- based reagent | rx1 3’HF f | this paper | with BamHI restriction site: GGCGGATCCCATTTATCAC TCGATACCATGAGCA | |
| Sequence- based reagent | rx1 3’HF r | this paper | with KpnI restriction site: GGCGGTACCTTCCAGTTTA AGAACATCCCCTCT | |
| Sequence- based reagent | rx1 sgRNA1 f | this paper | substituted nucleotides to facilitate T7 in vitro transcription of the sgRNA oligonucleotides are shown in small letters TAggAAATGCATGAGAGCGTCT | |
| Sequence- based reagent | rx1 sgRNA1 r | this paper | AAACAGACGCTCTCATGCATTT | |
| Sequence- based reagent | rx1 sgRNA2 f | this paper | substituted nucleotides to facilitate T7 in vitro transcription of the sgRNA oligonucleotides are shown in small letters TAggCTCTCATGCATTTATCAC | |
| Sequence- based reagent | rx1 sgRNA2 r | this paper | AAACGTGATAAATGCATGAGAG | |
| Sequence- based reagent | rx1 Lf | this paper | CTTTGCTGTTTTGAGAATTGCACC | |
| Sequence- based reagent | rx1 Lr | this paper | GAGACCGAACGATGACAATAACAC | |
| Sequence- based reagent | pDest f (control) | this paper | CGAGCGCAGCGAGTCAGTGAG | |
| Sequence- based reagent | pDest r (control) | this paper | CATGTAATACGACTCACTATAG | |
| Sequence- based reagent | pDest f mod | this paper | Asterisks indicate phosphorothioate bonds, ‘5’moiety’ was either 5’Biotin, Amino-dT or Spacer C3. 5’moiety-C*G*A*G*C*GCAGCGAGTCAGTGAG | |
| Sequence- based reagent | pDest r mod | this paper | Asterisks indicate phosphorothioate bonds, ‘5’moiety’ was either 5’Biotin , Amino-dT or Spacer C3. 5’moiety-C*A*T*G*T*AATACGACTCACTATAG | |
| Sequence- based reagent | pCS2 f | this paper | CCATTCAGGCTGCGCAACTG | |
| Sequence- based reagent | pCS2 r | this paper | CACACAGGAAACAGCTATGAC | |
| Sequence -based reagent | pCS2 f mod | this paper | Asterisks indicate phosphorothioate bonds, ‘5’moiety’ was either 5’Biotin, Amino-dT or Spacer C3. 5’moiety-C*C*A*T*T*CAGGCTG CGCAACTG | |
| Sequence- based reagent | pCS2 r mod | this paper | Asterisks indicate phosphorothioate bonds, ‘5’moiety’ was either 5’Biotin, Amino-dT or Spacer C3. 5’moiety-C*A*C*A*C*AGGAAACAGCTATGAC | |
| Sequence- based reagent | Gf | this paper | ATGGCAAGCTGACCCTGAAGTTCAT CTGCACCACCGGCAAGC | |
| Sequence- based reagent | Gr | this paper | CTCAGGTAGTGGTTGTCG | |
| Sequence- based reagent | gfpf | this paper | GCTCGACCAGGATGGGCA | |
| Sequence- based reagent | gfpr | this paper | CTGAGCAAAGACCCCAACGAGA AGCGCGATCACATG | |
| Sequence- based reagent | gfp probe f | this paper | GTGAGCAAGGGCGAGGAGCT | |
| Sequence- based reagent | gfp probe r | this paper | CTTGTACAGCTCGTCCATG |
Additional files
-
Supplementary file 1
Analysis of injected embryos.
Embryos injected with unmodified or modified (5’Biotin, Amino-dT, Spacer C3) long dsDNA gfp donor cassettes matching the rx2, actb, dnmt1 or rx1 locus, were scored for GFP expression and survival. Injections without Cas9 mRNA for control.
- https://doi.org/10.7554/eLife.39468.012
-
Supplementary file 2
Oligonucleotides used in this work.
Restriction enzyme sites used for cloning of PCR amplicons are indicated in italics. Substituted nucleotides to facilitate T7 in vitro transcription of the sgRNA oligonucleotides are shown in small letters (Stemmer et al., 2015). Locus primers forward (Lf) and reverse (Lr) of respective gene loci, gfp primers forward (Gf) and reverse (Gr), gfp sequencing primers gfpf and gfpr and primers to amplify the mgfp-flexible linker, as well as the gfp probe for Southern Blot analysis, are given. Asterisks indicate phosphorothioate bonds, ‘5’moiety’ was either 5’Biotin, Amino-dT or Spacer C3 in the pDest f mod, pDest r mod, pCS2 f mod and pCS2 r mod primers.
- https://doi.org/10.7554/eLife.39468.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39468.014





