Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells
Figures
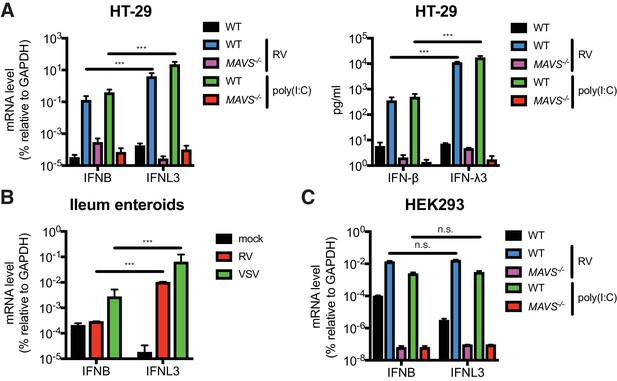
IECs predominantly produce type III IFN in response to RNA PAMP.
(A) WT and MAVS-/- HT-29 cells were stimulated with low molecular weight (LMW) poly (I:C) (200 ng/ml) or infected with the simian RV RRV strain (MOI = 1) for 24 hr. Expression of IFN-β and IFN-λ mRNA was measured by RT-qPCR and normalized to GAPDH expression. The supernatant of infected cells was harvested and IFN protein secretion was measured by ELISA. Clonal MAVS knockout was confirmed by western blot and Sanger sequencing. See Figure 1—figure supplement 1 for genomic sequences and Supplementary file 2 for sgRNA sequences. (B) Human intestinal enteroids were infected with VSV-GFP (MOI = 5) or the human RV WI61 strain (MOI = 5) for 16 hr. Expression of IFN-β and IFN-λ expression was measured by RT-qPCR and normalized to that of GAPDH. (C) WT and MAVS-/- HEK293 cells were stimulated with LMW poly (I:C) (200 ng/ml) or infected with RRV (MOI = 1) for 24 hr. Expression of IFN-β and IFN-λ expression was measured by RT-qPCR and normalized to that of GAPDH. For all figures, experiments were repeated at least three times with similar results. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).
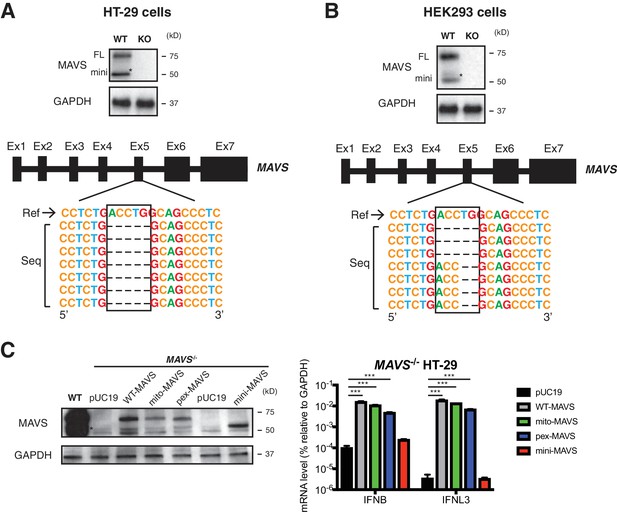
Construction and reconstitution of CRISPR-Cas9 MAVS knockout cell lines.
(A) Deletion validation of clonal MAVS-/- HT-29 (KO) cells by western blot and genomic sequencing. (B) Deletion validation of clonal MAVS-/- HEK293 (KO) cells by western blot and genomic sequencing. (C) WT and MAVS-/- HEK293 cells were transfected with WT or indicated MAVS mutants for 48 hr and harvested for western blot (left panel) or RT-qPCR analysis (right panel) measuring IFN-β and IFN-λ expression. For all figures except the lower panels of (a) and (b), experiments were repeated at least three times with similar results. Sanger sequencing was performed once for HT-29 and HEK293 cells. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001). (* represents mini-MAVS in all western blots).
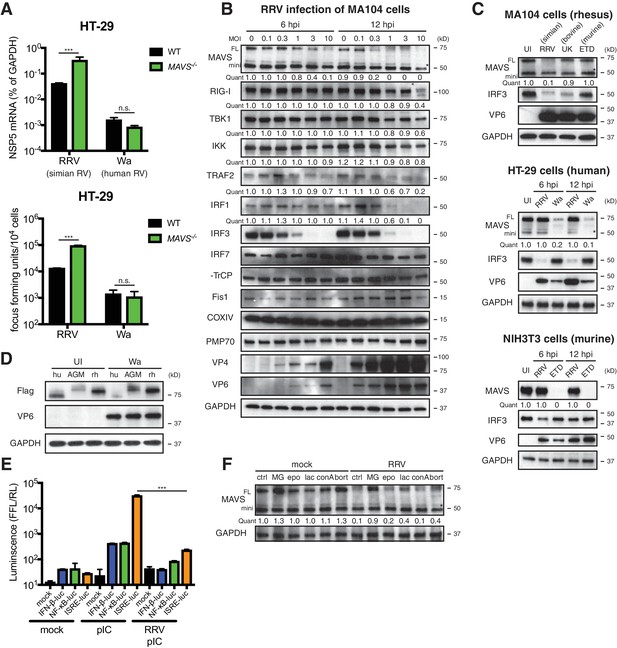
RV infection induces MAVS degradation in a host range restricted manner.
(A) WT and MAVS-/- HT-29 cells were infected with the simian RV RRV strain or the human RV Wa strain (MOI = 1) for 24 hr. Expression of viral gene NSP5 was measured by RT-qPCR and normalized to that of GAPDH. The virus yield in the supernatant was harvested and measured by a standard focus forming unit assay. (B) MA104 cells were infected with RRV at indicated MOIs for 6 or 12 hr. The lysates were harvested for western blot to examine the expression level of full-length (FL) and truncated mini-MAVS, other indicated RIG-I signaling factors, RV proteins (VP4, VP6) and GAPDH. The relative levels of individual proteins were quantified (quant) with respect to the GAPDH levels (lane 1 set as 1.0). (C) Rhesus MA104 cells, human HT-29 cells and murine NIH3T3 cells were infected with simian RRV, human Wa or murine ETD (MOI = 5) respectively and the protein levels of MAVS, VP6 and IRF3 were measured by western blot (UI: uninfected). (D) HEK293 cells were transfected with Flag-tagged MAVS from Homo sapiens (human, hu), Chlorocebus aethiops (African Green monkey, AGM), Macaca mulatta (rhesus monkey, rh) for 12 hr and infected with Wa (MOI = 3) for 12 hr. The protein levels of MAVS, VP6 and GAPDH were measured by western blot. (E) MA104 cells were transfected with luciferase expression constructs driven by IFN-β, NF-κB, or ISRE promoters, mock or infected with RRV (MOI = 3) for 8 hr, and stimulated with LMW poly (I:C) (pIC) for 8 hr. The level of firefly luciferase (FFL) was measured and normalized to that of renilla luciferase (RL), which serves as internal control. (F) MA104 cells were infected with RRV (MOI = 3) for 12 hr and treated with indicated proteasome and lysosome inhibitors for 12 hr. The lysates were harvested and MAVS level was measured by western blot. (MG: MG132; epo: epoxomicin; lac: lactacystin; conA: concanamycin A; bort: bortezomib). For all figures, experiments were repeated at least three times with similar results. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001). (* represents mini-MAVS in all western blots).
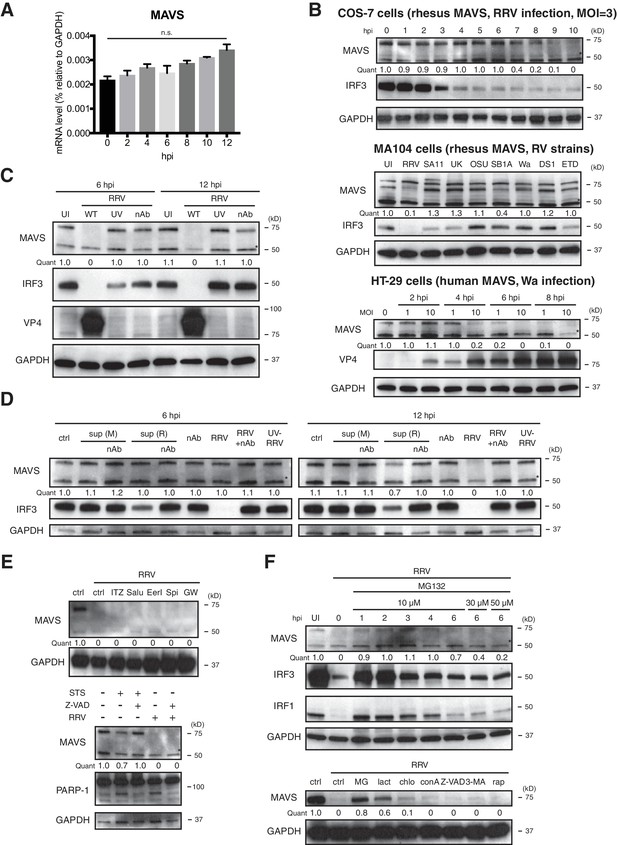
Active RV replication induces proteasomal degradation of MAVS.
(A) MA104 cells were infected with RRV (MOI = 1) and harvested at indicated time points post infection for RT-qPCR analysis. MAVS mRNA levels were measured and normalized to that of GAPDH. (B) African green monkey COS-7 cells were infected with RRV (MOI = 3) and harvested at indicated time points for western blot (top panel). MA104 cells were infected with different RV strains (MOI = 3) and harvested at 8 hpi for western blot (middle panel). HT-29 cells were infected with Wa at MOI = 1 or 10 for indicated time points and harvested for western blot. (RRV, SA11: simian RVs; UK: bovine RV; OSU, SB1A: porcine RVs; Wa, DS1: human RVs; ETD: murine RV). (C) MA104 cells were infected with RRV (MOI = 3), with RRV mixed with neutralizing antibodies (nAb) or with psoralen UV-inactivated RRV (UV) for 6 or 12 hpi. The lysates were harvested and examined by western blot. (D) Supernatants were harvested from mock-infected MA104 cells (sup(M)) or RRV-infected MA104 cells (sup(R)) and used to re-infect fresh MA104 cells. Lysates were harvested at 6 or 12 hpi and examined by western blot. (E) MA104 cells were infected with RRV (MOI = 3) for 12 hpi in the presence of different chemical inhibitors. (ITZ: itraconazole; Salu: salubrinal; EerI: Eeyarestatin I; Spi: spiroepoxide; GW: GW4869; STS: staurosporine; Z-VAD: Z-VAD-FMK). (F) MA104 cells were infected with RRV (MOI = 3) and treated with MG132 at indicated time point post infection (top panel). MA104 cells were infected with RRV (MOI = 3) for 12 hpi in the presence of different chemical inhibitors. (MG: MG132; lact: lactacystin; chlo: chloroquine; conA: concanamycin A; Z-VAD: Z-VAD-FMK; 3-MA: 3-Methyladenine; rap: rapamycin). For all figures, experiments were repeated at least three times with similar results. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).
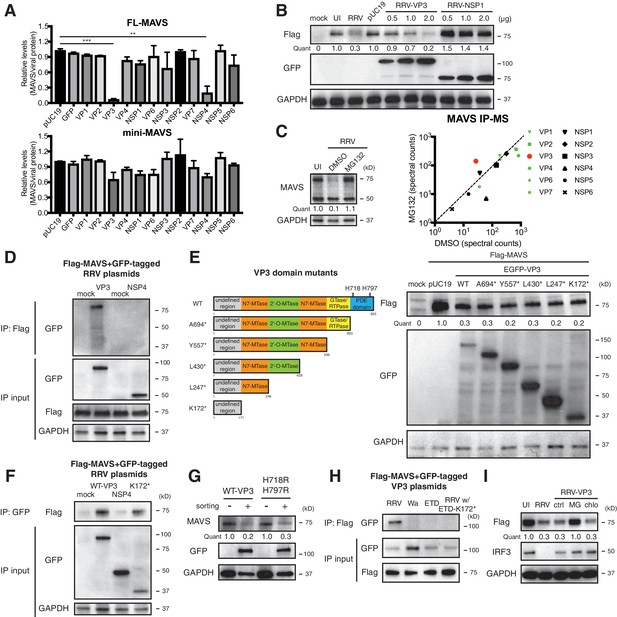
VP3 mediates MAVS interaction and degradation using an N-terminal domain.
(A) MA104 cells were transfected with plasmids expressing each of the 12 GFP-tagged RRV proteins for 72 hr and then subjected to sorting for GFP positive and negative cells. The levels of endogenous full-length and mini-MAVS were normalized to those of GFP-conjugated RRV proteins. (B) MA104 cells were co-transfected with 0.5 μg of Flag-tagged rhesus MAVS and GFP-tagged RRV-VP3 or RRV-NSP1 plasmids for 48 hr. RRV infection (MOI = 1) for 12 hr serves as a positive control. Total cell lysates were harvested and examined by western blot by indicated antibodies. (C) MA104 cells were infected with RRV (MOI = 3) for 8 hr in vehicle control (DMSO) or MG132 (10 μM) treatment. The lysates were subjected to immunoprecipitation using anti-MAVS antibody and analyzed by mass spectrometry for viral proteins. (D) MA104 cells were co-transfected with Flag-tagged MAVS and GFP-tagged RRV-VP3 or RRV-NSP4 for 48 hr and lysates were precipitated with anti-Flag antibody and GFP levels were examined by western blot. (E) Schematic diagram of WT and mutant VP3 proteins with defined domains illustrated in colors and catalytic sites of phosphodiesterase (PDE) activity indicated (left panel). MA104 cells were co-transfected with Flag-tagged MAVS and GFP-tagged VP3 mutants for 48 hr. The lysates were harvested and the levels of Flag and GFP were measured by western blot. (F) MA104 cells were co-transfected with Flag-tagged MAVS and GFP-tagged RV proteins (WT VP3, N-terminal K172* VP3, and NSP4) for 48 hr and lysates were harvested for immunoprecipitation using anti-GFP antibody. (G) MA104 cells were transfected with GFP-tagged wild-type or PDE mutant RRV-VP3 for 72 hr and then subjected to sorting for GFP positive cells. The levels of endogenous MAVS and GFP were examined by western blot. (H) MA104 cells were co-transfected with Flag-tagged MAVS and GFP-tagged VP3 from RRV (simian), Wa (human), ETD (murine) RV strains or chimeric RRV VP3 with 171 amino acids from ETD VP3, treated with MG132. The lysates were harvested for immunoprecipitation using anti-Flag antibody and probed for GFP levels. (I) MA104 cells were co-transfected with Flag-tagged MAVS and GFP-tagged RRV-VP3, and treated with MG132 (MG) or chloroquine (chlo) for 12 hr. The levels of Flag and IRF3 were examined by western blot. For all figures except (c), experiments were repeated at least three times with similar results. Experiments in (c) were performed once. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).
-
Figure 3—source data 1
Source data for Figure 3C: MAVS-RV protein IP counts.
- https://doi.org/10.7554/eLife.39494.008
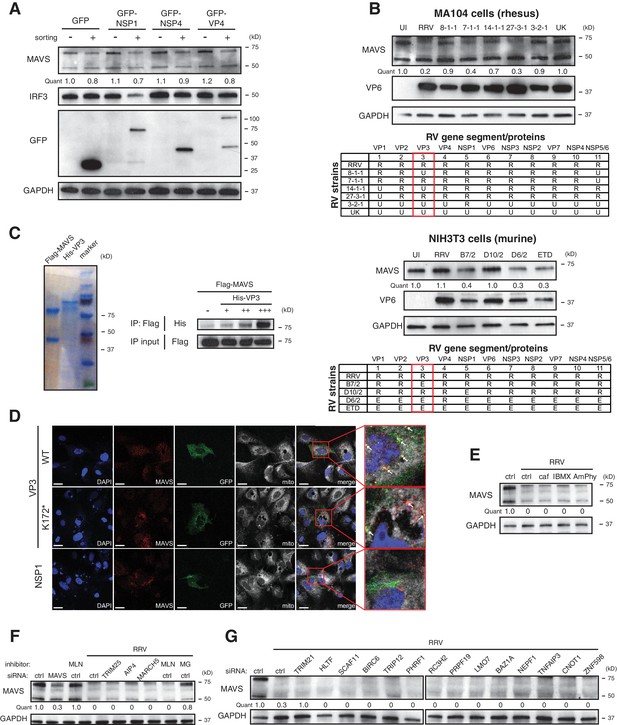
VP3 localizes to the mitochondria and induces MAVS degradation.
(A) MA104 cells were transfected with GFP-tagged RRV NSP1, NSP4 or VP4 for 72 hr and subject to sorting for GFP positive and negative cells. The levels of MAVS, IRF3, GFP and GAPDH were determined by western blot. Note that RV VP4 is cleaved into N-terminal VP8* and C-terminal VP5*. GFP tags are constructed on the amino terminus, hence giving the larger band (GFP-VP4) and the smaller band (GFP-VP8*). (B) MA104 cells were infected with reassortant RVs between simian RRV and bovine UK strains (MOI = 3) for 12 hr (top panel). NIH3T3 cells were infected with reassortant RVs between simian RRV and murine ETD strains (MOI = 3) for 12 hr (bottom panel). The levels of MAVS, VP6 and GAPDH were measured for both experiments. The genetic reassortment in different RV strains was illustrated in the table (R: RRV; E: EW). The red box highlights the RV gene three that encodes viral protein VP3. (C) Recombinant Flag-tagged MAVS and His-tagged VP3 proteins were purified from CHO-S cell lines and examined by Coomassie staining. Immunoprecipitation was performed using purified Flag-MAVS and an increase dose of His-VP3 with an anti-Flag antibody. The pull-down lysates and IP input were examined by western blot using anti-Flag and anti-His antibodies. (D) MA104 cells were transfected with GFP-tagged RRV VP3 (WT or N-terminal mutant K172*) or RRV NSP1 for 48 hr. The localization of MAVS and viral proteins were examined by confocal microscopy. Insets are enlarged by red boxes and co-localization of MAVS and viral protein is presented as yellow dots, as indicated by white arrows in the merged panel. Scale bar: 10 μm. (E) MA104 cells were infected with RRV (MOI = 3) for 12 hpi in the presence of phosphodiesterase inhibitors. (caf: caffeic acid; IBMX: 3-isobutyl-1-methylxanthine; AmPhy: aminophylline). (F) MA104 cells were transfected with siRNA against indicated E3 ubiquitin ligases for 48 hr and infected with RRV (MOI = 3) for 12 hr with or without inhibitor treatment (MLN: MLN4924; MG: MG132). The lysates were harvested and examined by western blot for the levels of MAVS and GAPDH. (G) MA104 cells were transfected with siRNA against indicated MAVS-interacting host proteins 48 hr and infected with RRV (MOI = 3) for 12 hr. The levels of MAVS and GAPDH were measured by western blot. For all figures, experiments were repeated at least three times.
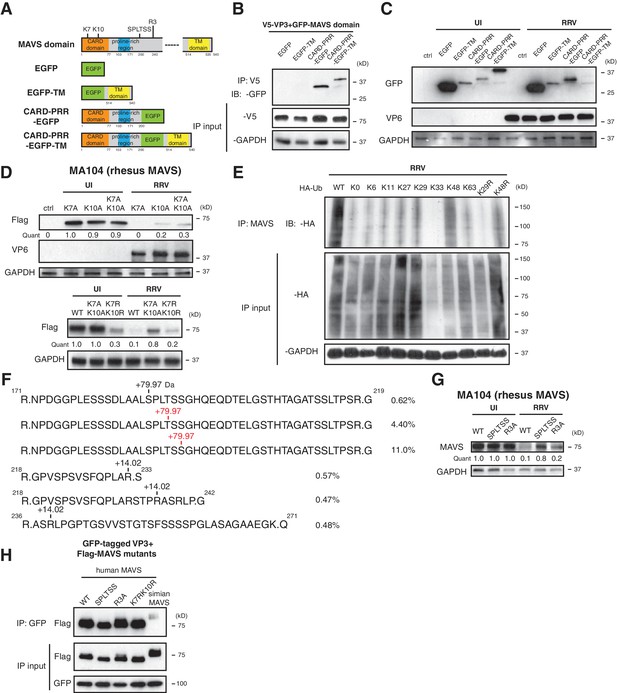
Phosphorylation mediates MAVS degradation during RV infection.
(A) Schematic diagram of MAVS and chimeric EGFP-MAVS proteins with defined domains illustrated in colors. Two lysines within the CARD domain and novel post-translational modifications (PTMs) are pointed out (left panel). (B) MA104 cells were transfected with indicated MAVS plasmids for 48 hr with or without RRV infection (MOI = 1) for the last 12 hr. The lysates were harvested and examined by western blot. (C) MA104 cells were co-transfected with GFP-tagged MAVS domain mutants and V5-tagged RRV VP3 for 48 hr and lysates were harvested for immunoprecipitation using anti-V5 antibody. (D) MA104 cells were transfected with indicated GFP-tagged MAVS mutants for 48 hr with or without RRV infection (MOI = 1) for the last 12 hr. The levels of MAVS and viral protein VP6 expression were measured by western blot. (E) MA104 cells were transfected with WT or indicated lysine-mutants of HA-tagged ubiquitin for 48 hr, infected with RRV (MOI = 1) for the last 12 hr and harvested for immunoprecipitation using an anti-MAVS antibody. The levels of ubiquitin-conjugated MAVS were measured using an anti-HA antibody. (F) Mass spectrometry analysis of PTMs on MAVS, immunoprecipitated from RRV-infected MA104 lysates at six hpi. Numbers indicate the mass increase (+79.97: phosphorylation;+14.02: methylation). The novel PTMs identified in this study are highlighted in red. The percentage of phosphorylation and methylation modification in total MAVS proteins is presented as %. (G) MA104 cells were transfected with WT or MAVS mutants (SPLTSS: SPLTSS motif mutated to six alanines; R3A: R232, R236, R239 mutated to three alanines) for 48 hr with or without RRV infection (MOI = 1) for the last 12 hr. The levels of MAVS were measured by western blot. (H) HEK293 cells were co-transfected with GFP-tagged Wa-VP3 and WT or MAVS mutants for 48 hr and treated with MG132. Lysates were harvested for immunoprecipitation using anti-GFP antibody and probed for MAVS levels. For all figures except (e) and (f), experiments were repeated at least three times with similar results. Experiment in (e) was performed twice and (f) was performed once.
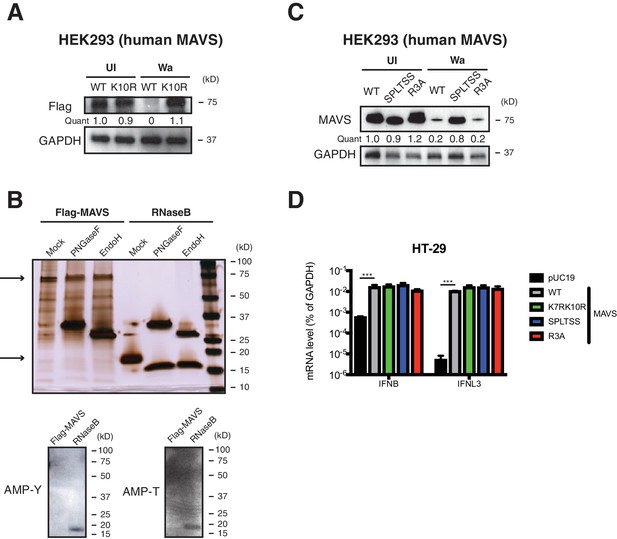
SPLTSS phosphorylation mediates MAVS degradation by RV infection.
(A) HEK293 cells were transfected with indicated Flag-tagged MAVS mutants for 48 hr with or without human RV Wa infection (MOI = 3) for the last 12 hr. The levels of MAVS and GAPDH were measured by western blot. (B) Purified recombinant Flag-MAVS and RNaseB proteins were digested with PNGaseF or EndoH and measured by silver staining (upper panel) and western blot for potential AMPylation (lower panel). Top arrow marks recombinant MAVS protein (~72 kD) and bottom arrow marks recombinant RNaseB protein (~17 kD). (C) HEK293 cells were transfected with indicated Flag-tagged MAVS mutants (SPLTSS: SPLTSS mutated to six alanines; R3A: R232, R236, R239 mutated to three alanines) for 48 hr with or without human RV Wa infection (MOI = 3) for the last 12 hr. The levels of MAVS and GAPDH were measured by western blot. (D) MAVS-/- HEK293 cells were transfected with WT or indicated MAVS mutants for 48 hr and harvested for RT-qPCR analysis measuring IFN-β and IFN-λ expression. For all figures, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).
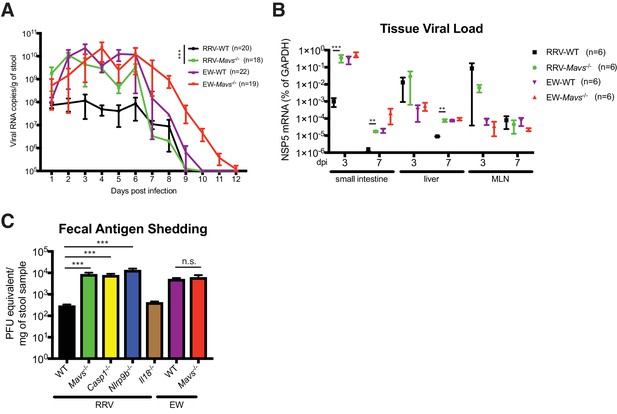
Replication of heterologous RV is enhanced in selected knockout mouse strains.
(A) Viral antigens in the fecal samples of EW or RRV-infected WT and Mavs-/- mice were measured by a standard ELISA assay over the course of 12 days. (B) Small intestines, liver and mesenteric lymph node (MLN) were harvested at 3 dpi or 7 dpi from EW or RRV infected animals and the levels of viral NSP5, indicative of virus replication, was measured by RT-qPCR and normalized to that of GAPDH. (C) C57Bl6/J mice with indicated gene deficiencies were infected with RRV (107 pfu) or EW (104 DD50). Three days later, feces samples were collected and tested by an ELISA assay for viral antigens and the amount of viruses were converted to PFU equivalents based on the standard curve. At least 8 mice were used in each group. For all figures, experiments were repeated at least two times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).
-
Figure 5—source data 1
Source data for Figure 5A.
- https://doi.org/10.7554/eLife.39494.013
-
Figure 5—source data 2
Source data for Figure 5B.
- https://doi.org/10.7554/eLife.39494.014
-
Figure 5—source data 3
Source data for Figure 5C.
- https://doi.org/10.7554/eLife.39494.015
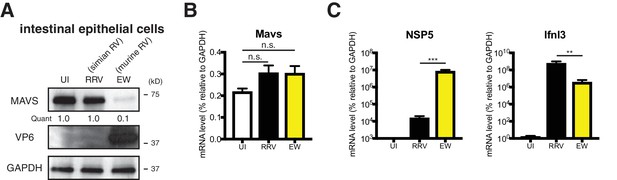
Heterologous RV induces robust type III IFN expression and is restricted by IFN and inflammasome signaling in vivo.
(A) 5 day old WT B6129SF2/J pups were infected with an equivalent dose of murine EW or simian RRV for 48 hr and small intestinal IECs were harvested using an EDTA-DTT isolation method. The levels of murine MAVS, RV protein VP6 and GAPDH were determined by western blot. (UI: uninfected). (B) 5 day old B6129SF2/J mice were infected with an equivalent dose of murine EW or simian RRV for 48 hr and IECs in the small intestine were harvested using an EDTA-DTT isolation method. The RNA levels of Mavs were measured by RT-qPCR analysis. (C) Same as (a) except that NSP5 and Ifnl3 were measured instead of Mavs. For all figures, experiments were repeated at least two times with at least four pups in each group. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*p≤0.05; **p≤0.01; ***p≤0.001).

Additional files
-
Supplementary file 1
Mass spectrometry data of MAVS immunoprecipitation in mock or RRV-infected MA104 lysates with or without MG132 treatment.
Units are colored in red based on the respective number of spectral counts. 12 rotaviral proteins are highlighted in yellow for easy identification. Column 1: Rank number based on the total number of spectral counts for all samples Column 2: Host (Chlorocebus genus) and viral proteins identified by mass spec Column 3: Raw spectral counts of uninfected MA104 sample Column 4: Raw spectral counts of RRV-infected MA104 sample Column 5: Raw spectral counts of RRV-infected, MG132-treated MA104 sample
- https://doi.org/10.7554/eLife.39494.016
-
Supplementary file 2
QPCR primer and siRNA information
- https://doi.org/10.7554/eLife.39494.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39494.018





