CRISPR knockouts reveal an endogenous role for ancient neuropeptides in regulating developmental timing in a sea anemone
Figures
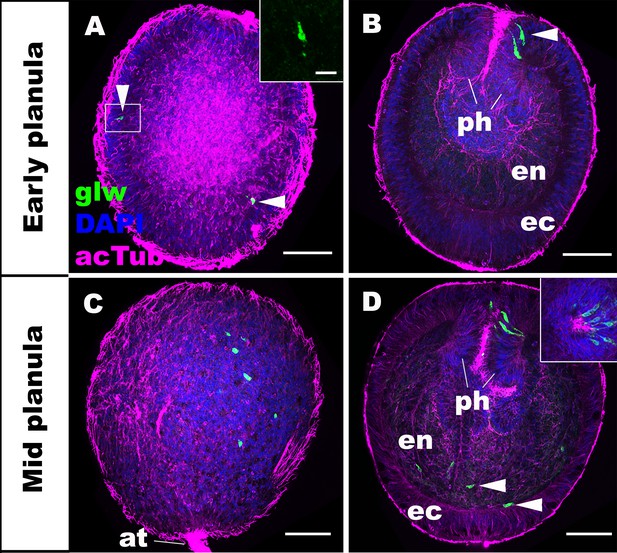
GLWamide precursor transcripts are expressed in ectodermal and endodermal epithelial cells of sea anemone planula larvae.
Z-projections of confocal sections of Nematostella vectensis at planula stages, labeled with an antisense riboprobe against GLWamide precursor transcript (‘glw’) and an antibody against acetylated ∂-tubulin (‘acTub’). Nuclei are labeled with DAPI. All panels show side views of animals with the blastopore/mouth facing up. The left columns (A and C) show superficial planes of section at the level of the ectoderm, and the right columns (B and D) show longitudinal sections through the center. (A, B) early planula. GLWamide transcripts are found in ectodermal sensory cells that are present in the outer epithelium and the pharynx (arrowheads in A and B). The inset in A show transcript-expressing epithelial cells in boxed regions with the apical side of the cell facing up. Note the spindle-shaped morphology characteristic of cnidarian sensory cells (Thomas and Edwards, 1991). (C, D) mid-planula. A subset of endodermal cells begin to express GLWamide transcript (arrowheads in D). The inset in D shows transverse sections of the pharynx, revealing the asymmetry in the spatial distribution of GLWamide-positive sensory cells. Abbreviations: ph pharynx; ec ectoderm; en endoderm; at apical tuft. Scale bar: 50 µm (A-D); 10 µm (inset in A).
-
Figure 1—source data 1
A confocal image stack used to generate panels A and B (dapi_blue actub_purple glw transcript_green).
- https://doi.org/10.7554/eLife.39742.004
-
Figure 1—source data 2
A confocal image stack used to generate panels C and D (dapi_blue actub_purple glw transcript_green).
- https://doi.org/10.7554/eLife.39742.005
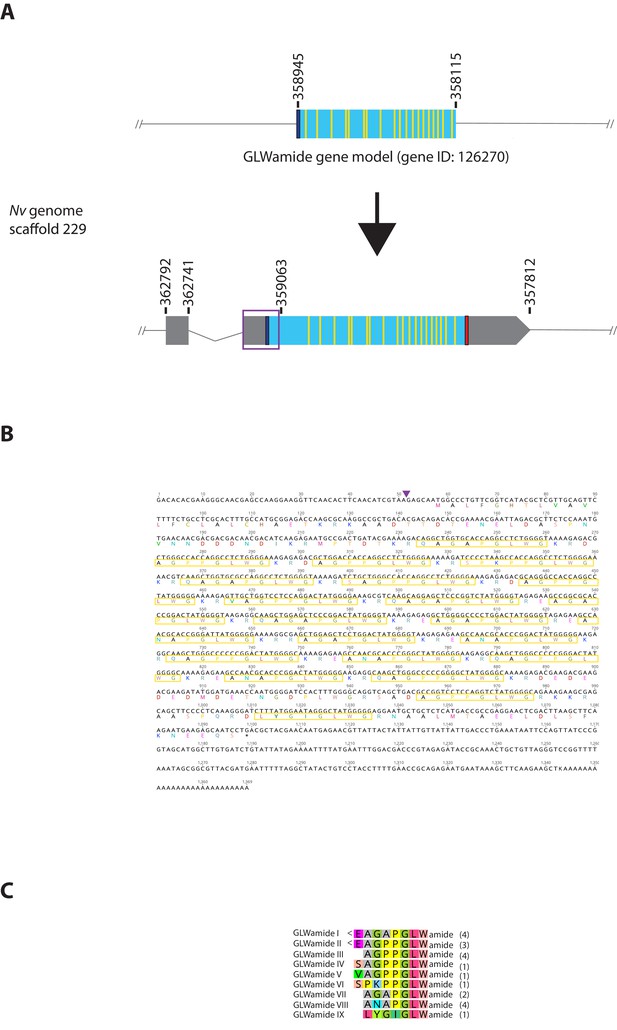
Genomic organization, cDNA sequences, and predicted mature peptides of GLWamide gene.
(A) Schematic view of the GLWamide genomic locus, located in scaffolds 229 of the N. vectensis genome (v.1.0; http://genome.jgi.doe.gov/Nemve1/Nemve1.home.html). Gene structure based on gene model prediction is shown at the top, and the revised structure based on the comparison of the N. vectensis genome and cDNA sequences is shown at the bottom. The number above the schematic indicate genomic coordinates within the scaffold. Grey boxes are untranslated regions; aqua-colored boxes are translated regions. Blue bars show predicted translation start sites; red bars show predicted translation termination sites; yellow bars show predicted GLWamide-encoding regions described below. The region boxed in purple is missing in the currently available genome of N. vectensis (v.1.0). Note that the revised gene structure contains two exons. (B) cDNA and deduced amino acid sequence of the GLWamide gene. Short peptides that are either repeated or similar, and are likely to be released by proteolytic cleavages are circled in yellow. It is assumed that single or multiple acidic and basic residues become cleaved as suggested in another sea anemone Anthopleura elegantissima (Darmer et al., 1991; Leviev and Grimmelikhuijzen, 1995). A purple arrowhead indicates the intron position. (C) Amino acid sequence alignment for GLWamides I-IX. We assume that upon release of the peptide, an N-terminal glutamine is spontaneously converted into a pyroglutamic acid residue (<Glu), and a C-terminal glycine becomes amidated by an α-amidating enzyme (cf. [Darmer et al., 1991]). The numbers on the right of each peptide sequence indicate copy numbers of each peptide that a single precursor protein is predicted to generate.
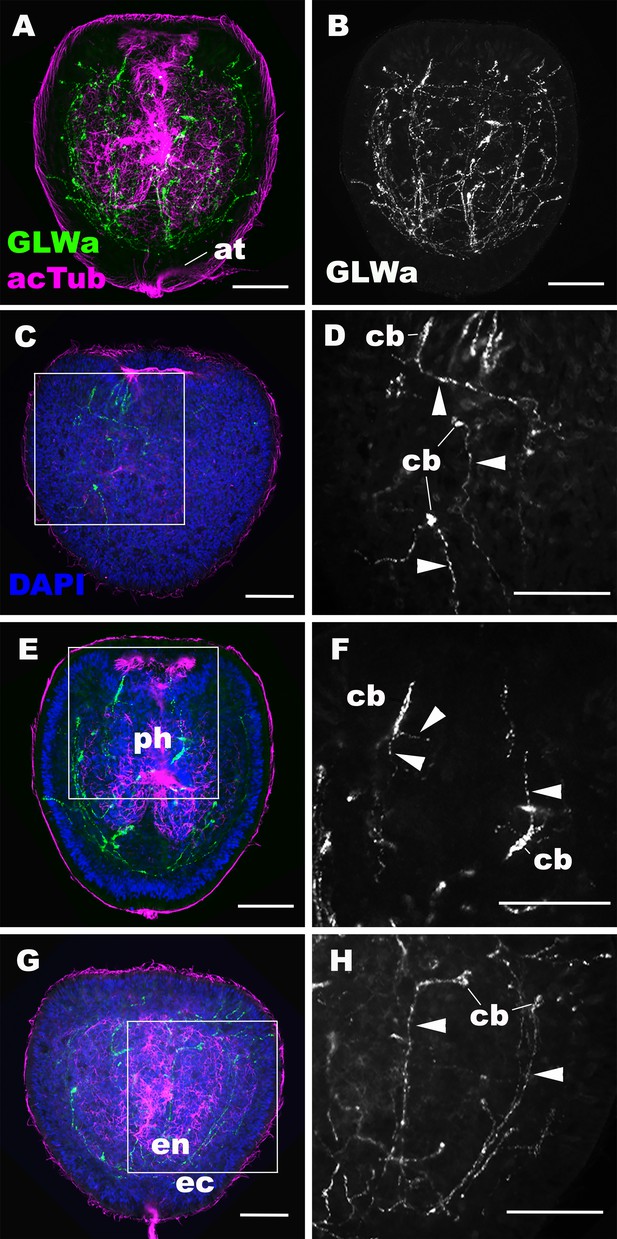
GLWamide-positive epithelial neurons form ectodermal and endodermal nervous systems of sea anemone planula larvae.
Z-projections of confocal sections of Nematostella vectensis at mid-late planula stages, labeled with antibodies against GLWamide (‘GLWa’) and acetylated ∂-tubulin (‘acTub’). Nuclei are labeled with DAPI. All panels show side views of animals with the blastopore/mouth facing up. (A and B) show sections through the entire animals. (C and D) show superficial planes of section at the level of the surface ectoderm. (E and F) show longitudinal sections through the center at the level of the pharyngeal sensory cells. (G and H) show longitudinal sections at the level of the endodermal neurons. Boxed regions in C, E and G are magnified in D, F and H, respectively. Note that ectodermal neurons in the surface ectoderm extend neurites laterally and aborally, but not orally (arrowheads in D), while pharyngeal ectodermal neurons extend neurites in all three directions (arrowheads in F). Endodermal neurons extend neurites that are part of longitudinally oriented neuronal bundles (arrowheads in H). Abbreviations: at apical tuft; cb cell body; ph pharynx; ec ectoderm; en endoderm. Scale bar: 50 µm.
-
Figure 2—source data 1
A confocal image stack used to generate panels A, B, E and F (dapi_cyan actub_green GLWa_red).
- https://doi.org/10.7554/eLife.39742.008
-
Figure 2—source data 2
A confocal image stack used to generate panels C, D, G and H (dapi_cyan actub_green GLWa_red).
- https://doi.org/10.7554/eLife.39742.009
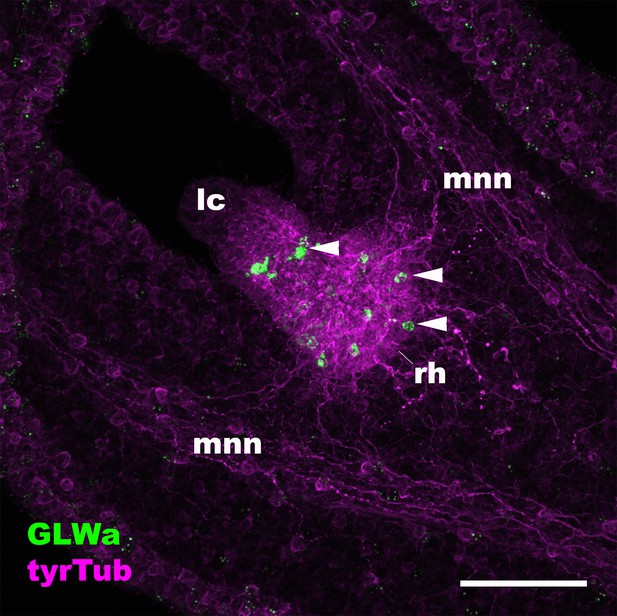
The anti-GLWamide antibody used in this study labels a subpopulation of ectodermal sensory cells of the rhopalial nervous system in the scyphozoan cnidarian Aurelia sp.1.
Confocal sections of a free-swimming ephyra of Aurelia sp.1, labeled with antibodies against GLWamide (‘GLWa’) and tyrosinated ∂-tubulin (‘tyrTub’). The panel shows an oral view of a rhopalium (rh) with its distal end (i.e. a lithocyst (lc)) pointing toward the upper left. Arrowheads show GLWamide-positive ectodermal sensory cells that are clustered in the proximal region of the rhopalium. This immunostaining pattern is consistent with that previously documented by using an antibody against EQPGLWamide (Nakanishi et al., 2009), indicating that the anti-GLWamide antibody used in this study can cross-react with GLWamides with divergent N-terminal sequences. Abbreviation: mnn motor nerve net. Scale bar: 50 µm.
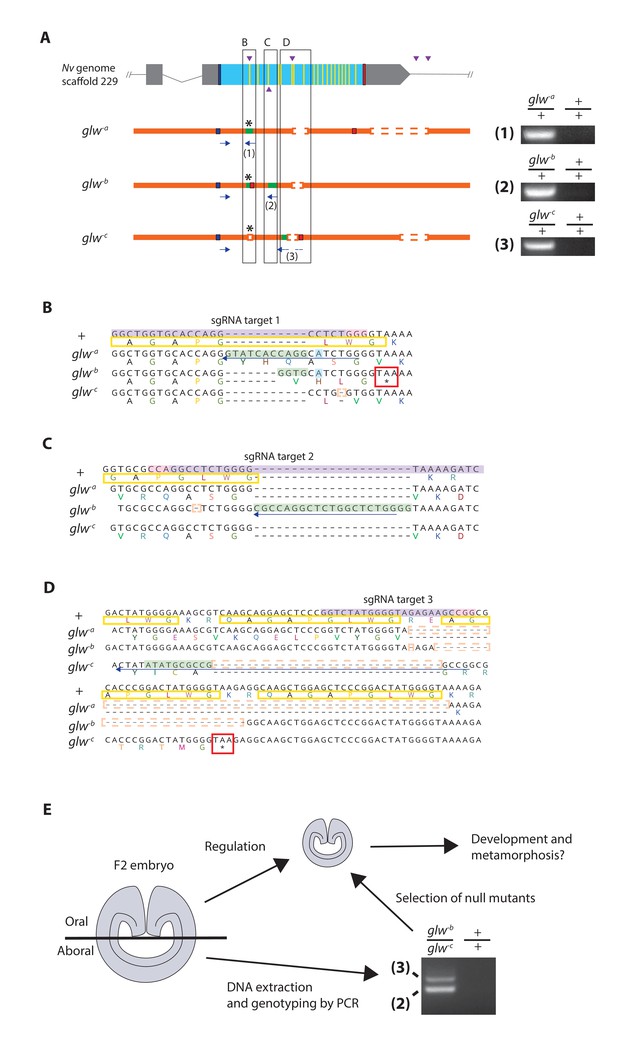
Generation of GLWamide null mutants by CRISPR-Cas9-mediated mutagenesis and F1 heterozygous mutant crosses.
(A) Schematic views of the GLWamide locus and mutant alleles (glw-a, glw-b and glw-c). Blue bars show predicted translation start sites; red bars show predicted translation termination sites; yellow bars show predicted GLWamide-encoding regions (Figure 1—figure supplement 1). Purple arrowheads show sgRNA target sites. Insertions are labeled in green, and deletions are shown blank. Asterisks show the sites of frameshift-causing mutations. Blue arrows mark regions targeted in the PCR analysis shown to the right; reverse primers are numbered (1 - 3). Note that mutant allele-specific primers (1 - 3) generate PCR products from genomic DNA samples of heterozygous mutants, but not from those of wildtype animals. (B-D) Nucleotide and translated amino acid sequences of wild-type and mutant alleles boxed in A. sgRNA target sites are shown in purple, and PAM sites are shown in pink. Predicted translation start sites are boxed in blue; predicted translation termination sites are boxed in red. Insertions are labeled in green, replacements in blue, and deletions boxed in dotted orange lines. Predicted immature neuropeptide sequences are boxed in yellow (cf. Figure 1—figure supplement 1). (E) a schematic showing the procedure of genotyping F2 embryos for developmental analyses. F2 embryos generated by F1 heterozygous mutant crosses were transversely bisected along the oral-aboral axis, and the oral halves were allowed to regulate and develop into polyps. Genomic DNA was extracted from single aboral halves and was used to genotype each embryo by PCR. The primers (2) and (3) were used to identify glw-b/glw-c embryos. '+' indicates a wildtype allele.
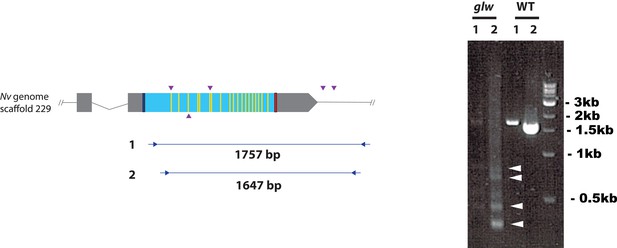
F0 embryos are mosaic mutants.
Schematic views of the GLWamide locus (left), and genomic DNA PCR results of an uninjected wildtype embryo (‘WT’) and an F0 embryo injected with locus-specific sgRNAs and Cas9 (‘glw’) (right). Blue bars show predicted translation start sites; red bars show predicted translation termination sites; yellow bars show predicted GLWamide-encoding regions as in Figure 2. Purple arrowheads show sgRNA target sites. Blue arrows mark regions targeted in the PCR analysis shown to the right. Note that genomic PCR of the WT embryo shows expected sizes of PCR fragments (1757 bp for primary PCR (‘1’), and 1647 bp for secondary nested PCR (‘2’)), while F0 embryos show additional bands of smaller sizes (arrowheads), indicating that targeted deletions of different sizes have occurred mosaically in each embryo. Primer sequences used for genomic PCR are: ‘1’ Forward 5’-GACACGACAGACACCGAAAACG-3’, ‘1’ Reverse 5’-GCATTTGGCAAATAAAACGCACATG-3’, ‘2’ Forward 5’-AATGTGAACAACGACGACGACAACG-3’, ‘2’ Reverse 5’-GGAGTGGTTTCCAAATCTCCCGAGC-3’.
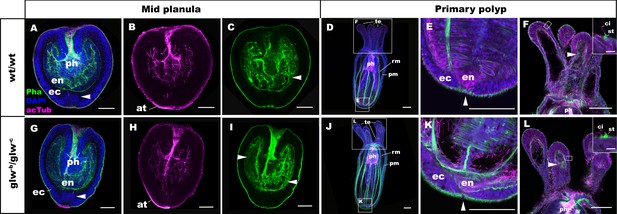
GLWamide null mutant planulae transform into morphologically normal primary polyps.
Confocal sections of wild-type (A-F) and mutant (glw-b/glw-c, G-L) Nematostella vectensis, labeled with an antibody against acetylated ∂-tubulin (acTub). Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI. All panels show side views of animals with the blastpore/mouth facing up. (A-C, G-I): mid-planula. Similar to the wild-type, glw-b/glw-c mutant planulae develop apical tufts (at) at the aboral pole; in addition, the basal translocation of nuclei in the aboral-most ectoderm (arrowheads in A, G, M), as well as the development of myofilaments in the endoderm (arrowheads in C, I), are evident. (D-F, J-L) primary polyp. glw-b/glw-c mutants develop into morphologically normal primary polyps, forming a set of four oral tentacles (D, J) with longitudinally oriented muscle fibers in the endoderm (arrowheads in F, L) and hair cells in the ectoderm--a polyp-specific cell type in N. vectensis (Nakanishi et al., 2012; Watson et al., 2009)-- characterized by a single cilium (ci) surrounded at the base by stereocilia (st) (insets in F, L). Note also the development of eight sets of longitudinally oriented parietal (pm) and retractor (rm) muscles in the endoderm of the body column (D, J), and the loss of apical tufts (arrowheads in E, K), in wild-type as well as glw-b/glw-c mutant animals. Abbreviations: ph pharynx; ec ectoderm; en endoderm. Scale bar: 50 µm; 5 µm (inset in F, L).
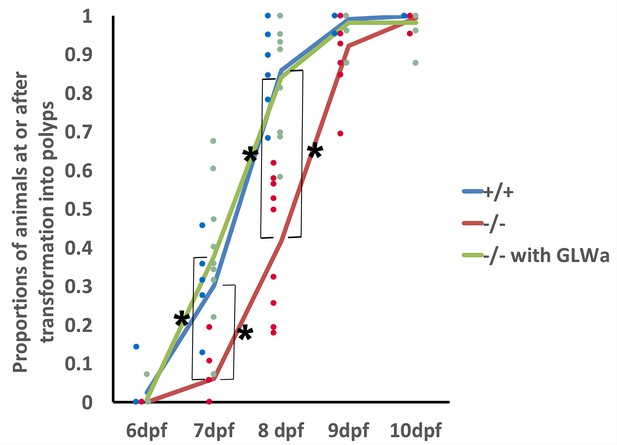
GLWamides regulate the timing of metamorphosis in sea anemone planula larvae.
A line graph showing changes in the proportions of animals at the tentacle-bud or primary polyp stage from 6 dpf to 10 dpf in F3 GLWamide null mutants (‘-/-‘), F3 wildtype control animals (‘+/+'), and F3 GLWamide null mutants treated with synthetic GLWamide peptides (‘-/- with GLWa’) at 16°C. The proportions of metamorphosing and metamorphosed animals were significantly lower in null mutants at 7 and 8 dpf relative to the wildtype controls and null mutants treated with GLWamide (one-tailed t-test: -/- relative to +/+, 7 dpf p<10^−3, 8 dpf p<10^−4; -/- relative to -/- with GLWa, 7 dpf p<10^−3, 8 dpf, p<10^−4). Total numbers of experiments: -/- n = 9; +/+ n = 6; -/- with GLWa n = 9. Filled circles represent data points (+/+ in blue, -/- in red and -/- with GLWa in green). * denotes statistically significant difference (α = 0.05).
-
Figure 5—source data 1
Numerical data represented in Figure 5.
- https://doi.org/10.7554/eLife.39742.015
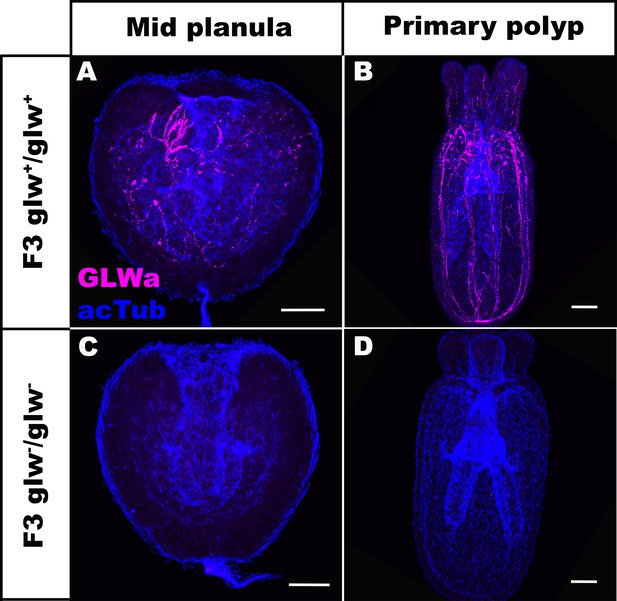
F3 GLWamide null mutant planulae transform into polyps.
Confocal sections of F3 wild-type (A, B) and mutant (glw-/glw-, C, D) Nematostella vectensis, labeled with antibodies against GLWamide (‘GLWa’) and acetylated ∂-tubulin (‘acTub’). All panels show side views of animals with the blastpore/mouth facing up. (A, C) Mid-planula. (B, D) Primary polyp. All panels show Z-projections of sections through the entire animals. F3 null mutants were generated by crossing F2 glw-a/glw-c adults with each other, and F3 wildtype animals were generated by crossing F2 glw+/glw+ adults with each other; glw-a/glw-c and glw+/glw+ F2 adults were siblings. Possible genotypes of F3 glw-/glw- null mutants were therefore glw-a/glw-a, glw-a/glw-c, or glw-c/glw-c. Note the lack of GLWamide immunostaining in glw-/glw- null mutants, which provides experimental evidence that there are no other GLWamide-like peptides present at the developmental stages investigated. Thus NvLWamide-like does not generate GLWamides, as predicted from the amino acid sequence data (see main text). Scale bar: 50 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (Nematostella vectensis) | F2 glw-a/glw-c | this paper | Nv F2 glw-a/glw-c_ this paper | See the Results section of the paper for a description of the biological sample. |
| Antibody | Anti-GLWamide (rabbit) | this paper | Anti-GLWamide_this paper:AB_2737386 | (1:200) See the Materials and Methods section of the paper for a description of the antibody. |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39742.016





