The mlpt/Ubr3/Svb module comprises an ancient developmental switch for embryonic patterning
Figures
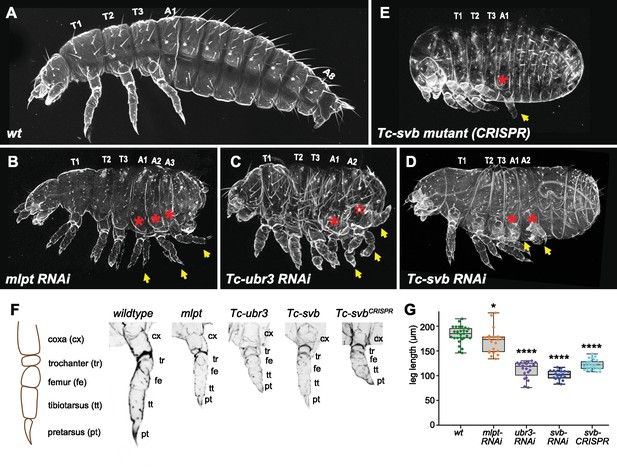
A cooperative segmentation function of the module Mlpt/Svb/Ubr3 in Tribolium.
Cuticle phenotypes of Tribolium first instar larvae from following genotypes: wild type (A), mlpt RNAi (B), Tc-ubr3 RNAi (C), Tc-svb RNAi (D), and Tc-svb CRISPR mutant (E). Depletion of mlpt, Tc-svb, and Tc-ubr3 causes highly similar segmentation phenotypes, characterized by a reduction in segment number, the presence of extra-legs (arrows) suggestive of transformation of abdominal segments towards a thoracic fate (red asterisks), and the frequent absence of terminal structures. (F) Knockdown of each of the three genes leads to shortened ‘true-thoracic’ legs, with rounded and often poorly separated distal segments. The scheme represents a larval leg with corresponding segments; pictures portray an example of prothoracic leg (T1) in wildtype, mlpt, Tc-ubr3 and Tc-svb inactivation. (G) Quantification of the reduction in leg length, estimated by the distance between coxa/trochanter boundary to the pretarsus tip. Data were analyzed by one-way ANOVA using multiple comparison tests against wild-type values. *, p-value<0,05; ****, p-value<0,0001. Source data for Figure 1G are found in Source Data File 1.

Cuticle of Tribolium larvae showing different examples of Tc-ubr3 RNAi phenotypes.
(A–D) Knockdown larvae have strong segmentation phenotypes, with drastically antero-posteriorly shortened bodies. Posterior abdominal segments are missing and the remaining ones appear to be transformed to thoracic identity, with ectopic legs (yellow arrows) and/or spiracles resembling those normally present in T2 segments (white arrows). The leg-bearing transformed segments are marked by red asterisks. Legs are shortened and rounded, with reduced pretarsi. (E) A magnified image of the legs. Leg segments sometimes appear double- jointed (black arrow). coxa (c); trochanter (t); femur (f); tibiotarsus (tt); pretarsus (pt). (F) Magnified image of the head showing the shortened, rounded and bulbous head appendages in Tc-ubr3, when compared to wild type. Setae are usually missing on the antennae.
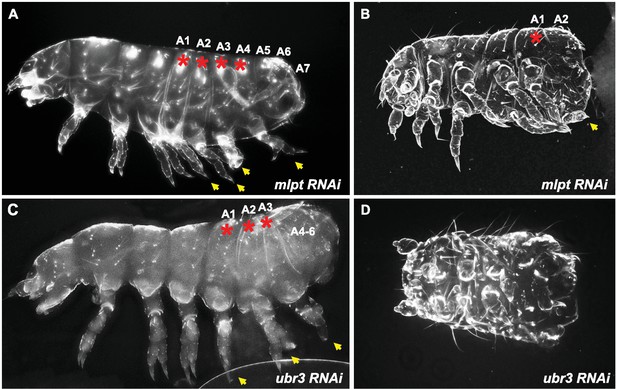
Extreme phenotypes of Tribolium mlpt and Tc-ubr3 knockdowns.
(A) A mlpt knockdown larva with four pairs of legs on transformed abdominal segments. The leg-bearing transformed segments are marked by red asterisks. (B) A mlpt knockdown with a strong segmentation phenotype, with one pair of ectopic legs and only two remaining abdominal segments. (C) A weak Tc-ubr3 knockdown phenotype, that resembles the mlpt phenotype in (A), with three pairs of ectopic legs and fused remaining abdominal segments. (D) A drastically shortened Tc-ubr3 knockdown larva, apparently lacking all abdominal segments.

Tribolium Tc-svb RNAi larval cuticles of increasing phenotypic strength.
(A–D) In larvae with weaker segmentation phenotypes, bodies are shortened along the anterior-posterior axis. While most of the abdominal segments are often present, segment boundaries are less distinct especially in the latter abdominal segments where they may be fused, and telson appendages are usually missing. Thoracic legs display shorter leg segments (See Figure 1). (E–G) Stronger phenotypes are more antero-posteriorly shortened and the last abdominal segment and the telson may either be absent or fused with the anterior segments beyond distinction. Leg segments get progressively shorter and more rounded, and the pretarsi reduced. Sometimes urogomphi (u) are present (G). Irrespective of body size and phenotypic strength, segments with extra legs are always A1 and A2 (red asterisks). One or more of these ectopic legs are often reduced to stumps. (H) Magnified ventral view of a svb knockdown showing transformed abdominal segments bearing legs (red asterisks) and fused abdominal segments.

Schematic representation of Tc-shavenbaby locus (A) and transcript (B), showing the site at which a GFP-containing marker plasmid was inserted by CRISPR/cas9 genome editing (see also Materials and methods).
The mutagenic cassette is inserted into exon 2, that is within the open reading frame upstream of the region encoding the DNA-binding zinc finger domain. Gene disruption leads to mRNA truncation after the insertion site, since Tc-svb expression is absent in homozygous mutants. In addition to segmentation defects and transformation toward thoracic identity, other phenotypes observed in Tc-svbCRISPR mutants include incipient spiracles (possibly a secondary effect of cuticle thinning leading to a defect in the development of tracheal rings); sensory bristles that are shorter and thicker; leg segment boundaries that are not clearly defined; missing leg bristles; unsclerotized pretarsi with soft, rounded apices; and antennae lacking the terminal setae. Therefore, late functions of Tc-svb in epidermal and appendage differentiation are strongly affected in Tc-svbCRISPR mutant embryos, while the segmentation phenotype is milder that Tc-svb-RNAi knockdown due to maternal contribution of Tc-svb (Ray et al., in preparation).
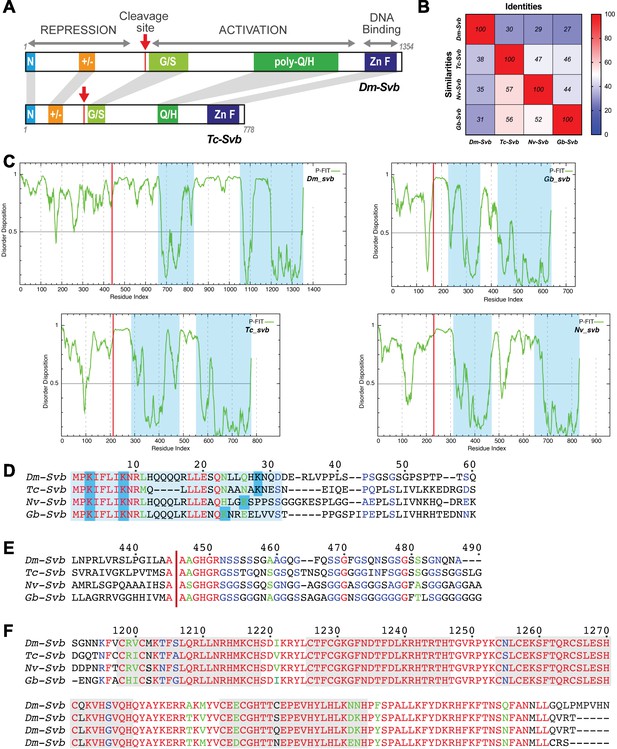
Shavenbaby protein features are conserved across insect species.
(A) Schematic representation of Drosophila (Dm-Svb) and Tribolium (Tc-Svb) proteins, highlighting conserved regions. These include the N-terminus (blue), stretches of charged amino acids in the repressor region (orange), Mlpt-dependent proteolytic maturation site (red arrow) followed by a glycine/serine-rich region (light green), poly-glutamine/histidine stretch (green) and the zinc finger DNA-binding domain (blue). (B) Sequence conservation of the Svb protein across insects. The heat map represents percentages of sequence identity and similarity. (C) Disorder disposition of each protein was evaluated using PONDR-FIT (http://www.disprot.org/pondr-fit.php). Svb proteins share intrinsic disorder disposition at their N-termini compared to their DNA binding and transactivation domains (blue shade). The red line indicates the maturation site in each protein. (D) Evolutionary conservation of the N-terminal degron, with three key lysine residues required for Ubr3-mediated processing of Svb. (E) A highly conserved AAGHGR motif flanks the maturation site (red arrow). (F) The zinc finger domain is also highly conserved across Svb proteins.
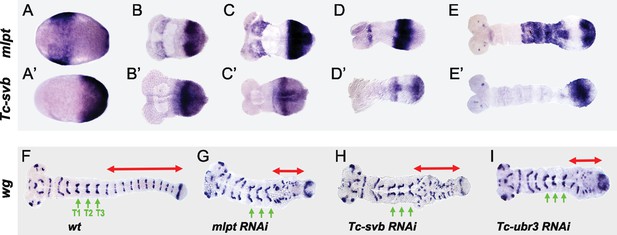
Tc-mlpt and Tc-Svb embryonic expression and function of the mlpt/svb/ubr3 module in abdominal patterning.
(A–E’) Whole mount in-situ hybridization of Tribolium embryo showing mRNA expression of mlpt and Tc-svb from late blastoderm (A,A’) through extending germband stages (B,B’, C,C’, D,D’, E,E’), highlighting their complementary expression pattern (F–I) Wingless (wg) expression in wild type (F), mlpt-RNAi (G), Tc-svb- RNAi (H) and Tc-ubr3- RNAi (I) Tribolium embryos. Abdominal segments are highlighted with red arrow. ln all three knockdown conditions, wg segmental stripes are disrupted right after the last (T3) thoracic stripe. Thoracic segments (T1–T3) are indicated by green arrows.
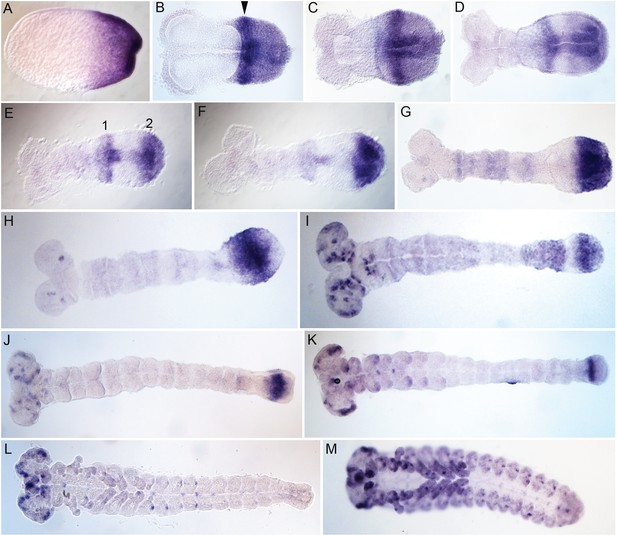
Embryonic expression of Tribolium svb.
Tc-svb is expressed at the posterior end in the blastoderm and primitive pit stage embryos (A). In the germ rudiment, the posterior expression assumes a composite pattern with strong expression in an anterior band abutting the serosa window (arrowhead) and diffuse expression posteriorly (B and C). In the early germband, the diffuse expression clears out anteriorly, and concentrates as a distinct domain at the posterior end giving rise to two distinct domains, 1 and 2 (D and E). Domain one gradually fades but remains spread out over a large area (F, G). Domain two detaches from the posterior end but remains close to it (H–K). Tc-svb also appears as dots in the head lobe in the extending germband (H) assuming a complex pattern in neuronal cells at the end of segmentation (I–L). Tc-svb is expressed in the gnathal and thoracic appendages as segmental dots, and as dots along the ventral midline, after the completion of segmentation (K and L). In old embryos, Tc-svb is strongly expressed in the presumptive antennae, gnathal and thoracic appendages, and on the pleuropods on the first abdominal segment (M).

mlpt RNAi causes de-repression of svb expression in short germ embryos of Tribolium and Oncopeltus.
(A,B) Wild-type expression of Tc-svb in Tribolium embryos. (A) Early germ rudiment expression of Tc-svb begins as a posterior cap, with a strong stripe of expression near the anterior boundary, and a more diffuse posterior expression. In the early to mid germband stages, Tc-svb expression fades somewhat anteriorly the diffuse posterior expression clears resolving into two distinct domains, a fading anterior and a newly emerging posterior (B). (A’,B’) Expression of Tc-svb in mlpt RNAi Tribolium embryos. (A’) Germ rudiment expression of Tc-svb in mlpt RNAi embryos keeps strong in the posterior and the anterior band of expression is lost. (B’) Early to extending germband stages in mlpt RNAi show a similar absence of the anterior band and a strong persistent posterior expression, significantly stronger than in wild type embryos, suggesting de-repression of Tc-svb expression due to reduced mlpt levels. (C–D) Wild-type expression of Of-svb in Oncopeltus embryos. (C) Early germband expression of Of-svb is restricted to the growth zone, where a strong central stripe and a fainter anterior stripe of expression can be seen. (D) Late germband expression of Of-svb is seen in a single strong domain of posterior growth zone expression, in the limb buds, and in neurons in the head lobes. (C’–D’) Expression of Of-svb in mlpt RNAi Oncopeltus embryos. Of-mlpt RNAi germband embryos display distal limb bud expression of Of-svb, and exhibit ectopic svb expression throughout the distorted germband, in the head, and throughout the growth zone (red arrows).
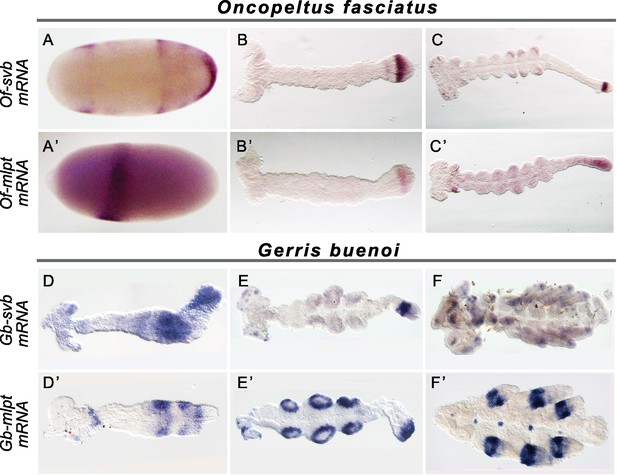
Expression of mlpt and svb in hemipteran embryos.
Whole mount in situ hybridization of svb and mlpt mRNA in Oncopeltus (A–C) and Gerris (D–F) embryos at early, mid-germ and late embryonic stages. (A–C) Oncopeltus embryonic expression. At early stages, Of-svb expression is mainly expressed in two domains (anterior head and thoracic segments) (A) Of-mlpt is restricted to a single strong stripe in presumptive head segments (A’). Then, Of-svb is expressed faintly in the head lobes and strongly in two growth zone stripes (B) while Of-mlpt is exclusively expressed in the posterior of the growth zone (B’). Late embryos express Of-svb expression in a strong stripe in the middle of the growth zone, as well as in putative head neurons and limb buds (C). At this stage, faint Of-mlpt mRNA expression is detected in the head appendages, putative head and thoracic segments, and strong but diffuse expression throughout the growth zone (C’). (D–F) Gerris embryonic expression. In early embryos, Gb-svb is faintly expressed in the head and thorax, with stronger expression in the abdomen of the early germ band (D), when Gb-mlpt expression is restricted to a thoracic stripe and two distinct abdominal domains, abutting Gb-svb expression (D’). Mid germ band embryos have more restricted Gb-svb expression, in a stripe in the growth zone, in putative neurons in the head, and faintly in limb buds (E) while they exhibit strong expression of Gb-mlpt in the limb buds, and in the posterior of the growth zone, immediately adjacent to strong Gb-svb expression. Late stage embryos exhibit faint banded expression of Gb-svb in the legs and head appendages, and in foci in the head (F) whereas they exhibit strong Gb-svb expression in the mature limbs, and in foci of expression along the embryo midline (F’).
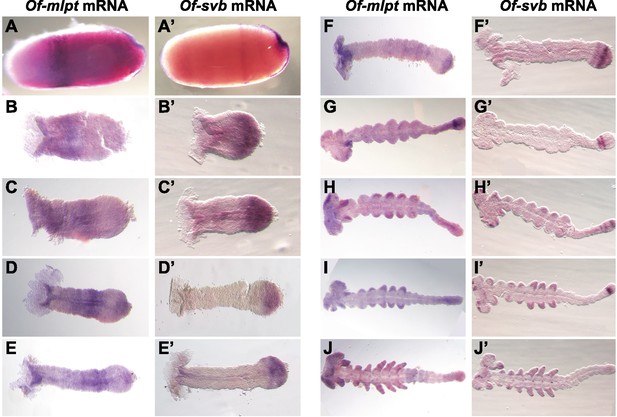
Expression of Of-mlpt and Of-svb mRNA throughout Oncopeltus embryogenesis.
(A–J) Expression of Of-mlpt. (A) Blastoderm expression of Of-mlpt includes an anterior stripe and emergent expression adjacent to, and underlying, the invagination at the onset of gastrulation. (B–E) Early germband embryos exhibit moderate staining in the head lobes, and a broadening anterior domain in thoracic segments, which moves posteriorly as the germband extends. Of-mlpt expression is also evident in the posterior growth zone (D). As the anterior domain fades (F), the posterior domain broadens and darkens. As germband growth continues (G–H), this domain moves anteriorly from the growth zone. Of-mlpt expression is seen in developing limb buds and in head appendages. Towards the end of embryogenesis, Of-mlpt is broadly expressed throughout the unsegmented posterior germband (H–I), and exhibits banded expression in the limbs and strong head staining (J). (A’–J’) Expression of Of-Svb. (A’) Blastoderm Of-svb is expressed in two broad domains at the anterior and posterior of the embryo, with a clearing in the blastoderm middle. Strong expression is seen at the invagination when gastrulation begins. (B’–E’) In the early germband, Of-svb mRNA is seen in a strong domain covering the entire embryo posterior, with low levels throughout the embryo anterior. This clears before limb bud formation (D’), when the posterior domain becomes restricted to the posterior of the growth zone (E’). Posterior expression matures into low level expression throughout the growth zone, with a strong stripe in the middle third adjacent to more posterior Of-mlpt expression (G’), and with low level expression overlapping Of-mlpt (F’). This stripe becomes two stripes that move anteriorly within the growth zone (G’) before emergence of a second growth zone stripe (H’), concomitant with expression in the nascent limb buds and in the head lobes in discrete spots; the unsegmented germband contains several concurrent stripes of svb expression (H’). The new, most posterior domain of Of-svb darkens during limb bud growth (I’), and eventually disappears at the end of germband elongation (J’).
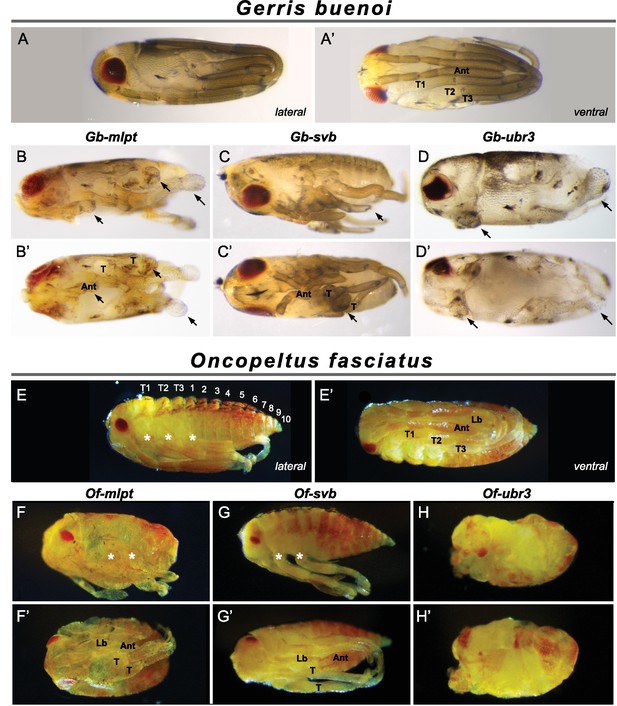
Knockdown of mlpt, svb, and ubr3 affects embryo segmentation in Gerris (A–D’) and Oncopeltus (E–H’).
Hatchlings are presented in lateral (A–D, E–H) and ventral (A’–D’ and E’–H’) views. Wild type Gerris pre-nymphs possess red pigmented eyes, and antennae that extend along the ventral side of the embryo, terminating between long legs which wrap around the embryo (A–A’). Both Gb-mlpt and Gb-svb RNAi embryos display posterior truncation, as well as loss and/or fusion of legs and head appendages (B–C’). Gb-mlpt embryos show altered eye morphology. Gb-ubr3 embryos exhibit more severe posterior, leg and eye phenotypes (D,D’). (E–H’) Phenotypes of wild type Oncopeltus (E–E’) hatchlings alongside Of-mlpt (F–F’), Of-svb (G–G) and Of-ubr3 (H–H’) RNAi. Of-mlpt and Of-svb RNAi causes posterior truncation, with the fusion/loss of thoracic segments, shortened legs and head appendages, and a reduced eye. Of-ubr3 RNAi displays similar phenotypes but stronger than Of-mlpt and Of-svb RNAi, with an apparent loss of axial polarity in severely affected Of-ubr3 RNAi embryos. Source data for Figure 4—figure supplements 1–3 are found in Source Data 1.
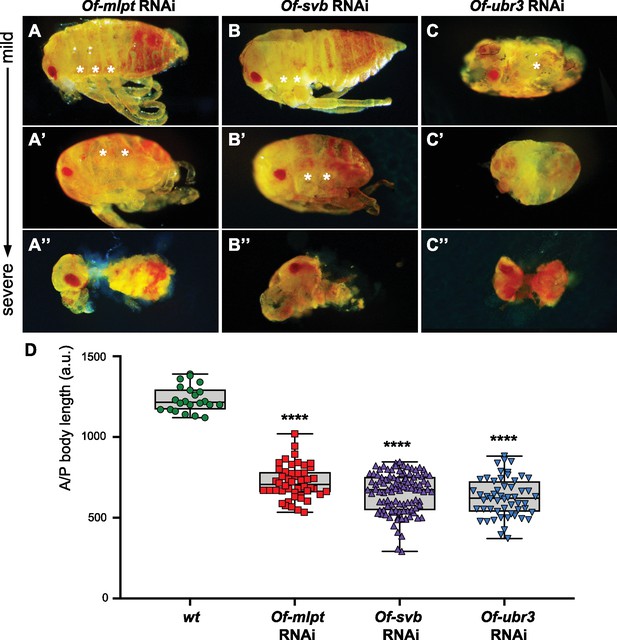
Phenotypes of increasing strength for Of-mlpt, Of-svb and Of-ubr3 RNAi in Oncopeltus.
(A–A”) Of-mlpt RNAi results in posterior truncation of the embryo. Mild phenotypes include abdominal shortening and weak limb defects (A), while moderate phenotypes cause strong abdominal truncation and thoracic segment fusions (A’). Severe Of-mlpt RNAi phenotypes result in loss of most abdominal segments, as well as thoracic and head appendages, and anterior segment loss (A’’). (B–B”) Weak phenotypes observed upon Of-svb RNAi knockdown include thoracic segment fusions and mild posterior abdominal segment loss (B); moderate phenotypes show severe posterior truncation, leg fusion and patterning defects (B’). Severe Of-svb RNAi phenotypes result in loss of most abdominal and thoracic segments and reduction of remaining head appendages (B”). Ofas ubr3 RNAi (C–C”) causes severe patterning defects. The mildest Of-ubr3 RNAi phenotypes, even at low dsRNA concentrations, result in loss of most abdominal segments and severe reduction or loss of leg segments (C). Moderate Of-ubr3 RNAi phenotypes cause further embryo reduction and appendage loss (C’), and severe Of-ubr3 RNAi phenotypes results in near ablation of the embryo; only limited ectodermal tissue remains, which may include a severely reduced head and loosely connected body segments of uncertain identity (C’’). (D) Quantification of Ofas RNAi embryo truncation phenotypes. Wild type (wt) as well as RNAi hatchlings from knockdown of each gene were measured from head to most distal point along the midline. Measurements were grouped for each genotype and compared using one-way ANOVA. ****, p-value<10E-9. N = 22 (wt), 48 (Of-mlpt), 106 (Of-svb) and 54 (Of-ubr3).
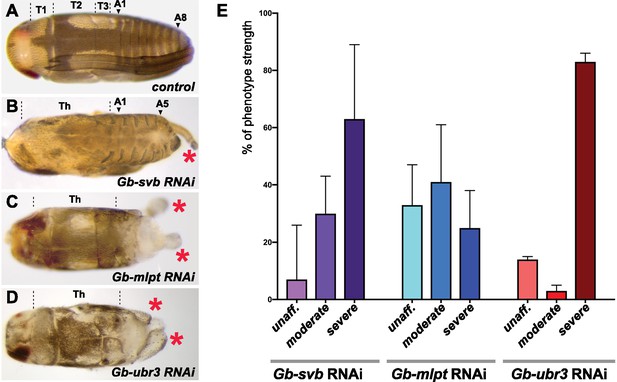
Effects of Gb-mlpt, Gb-svb, Gb-ubr3 RNAi depletion in Gerris.
Dorsal view of control (A), Gb-svb RNAi (B), Gb-mlpt RNAi (C) and Gb-ubr3 RNAi (D) Gerris buenoi prenymphs. (A) Morphological landmarks highlight the three thoracic (T1–T3) and abdominal (A1–A8) segments; the long T2 legs wrap around the dorsal surface in wild type. (B) A moderately affected Gb-svb RNAi hatching displays posterior truncation (segments posterior to A5 appear fused, if present). Note also strong defects in the dorsal midline. (C,D) Severe Gb-mlpt RNAi and Gb-ubr3 RNAi phenotypes, where most normal posterior structures are absent and/or fused. In all cases, RNAi embryos show alteration of thoracic appendages (red asterisks). (E) Frequency of phenotype strength observed following treatment with Gb-svb RNAi, Gb-mlpt RNAi and Gb-ubr3 RNAi. N = 146, 169 and 59 prenymphs, respectively.
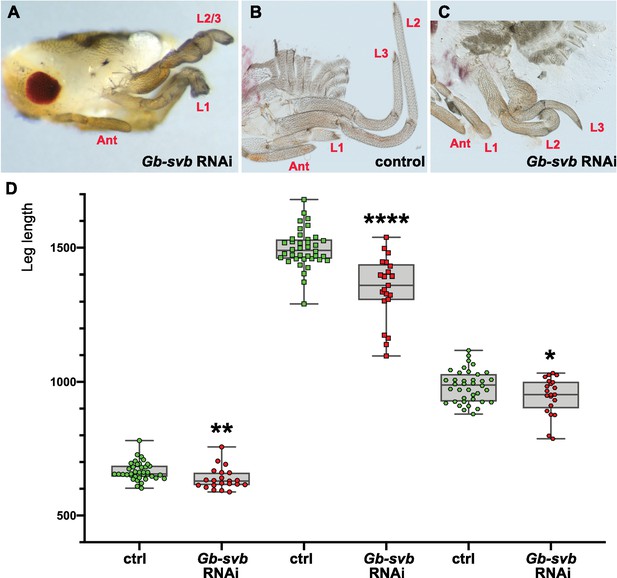
Gb-svb RNAi treatment induces defects in Gerris buenoi leg development and differentiation.
In most cases, legs are missing or fused (A). In mildest phenotypes where the three legs (L1–L3) remain individualized, they appear rounded and shorter, as seen on bisected control (YFP-RNAi, (B) and Gb-svb RNAi (C). (D) Quantification of individual leg length in the water strider Gerris. Twenty late embryos from females injected with Gb-svb ds-RNA and 22 embryos from females injected with YFP negative control were dissected and their legs measured. Gb-svb RNAi induces shortening of all legs. Data were analyzed using unpaired t tests. ****, p-value<0,0001; **, p-value<0,01; * p-value<0,05. Ant, antenna.
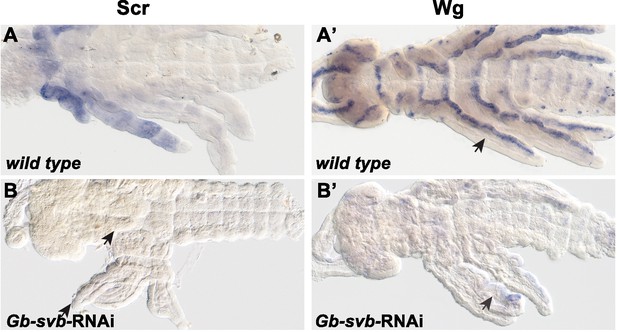
Gb-svb RNAi Gerris embryos display developmental defects.
Embryos show fusion of segments along the body axis as revealed by staining of sex-combs-reduced (Scr, (A, B), and Wingless (Wg, (A’, B’).
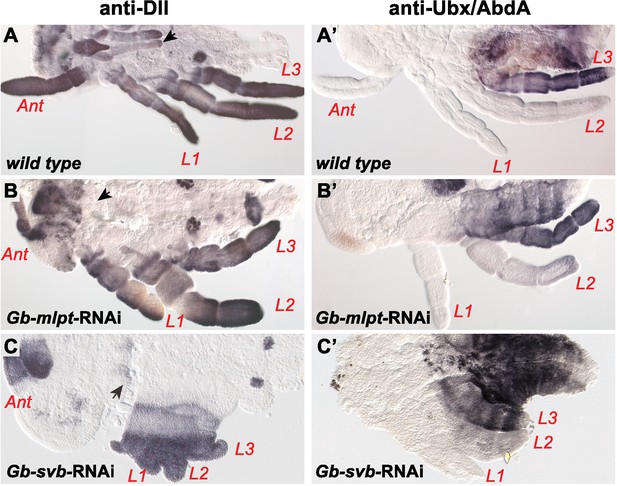
Effects of Gb-svb-RNAi and Gb-mlpt RNAI treatment in Gerris appendages.
Developmental defects in Gerris buenoi embryos following Gb-mlpt RNAi (B,B’) and Gb-svb RNAi (C,C’) treatment. Both RNAi affects legs, antennae and mouth parts (black arrows), as shown by immunostaining against Distal-less (Dll) (A, C) and Ultrabithorax/Abdominal A (Ubx/AbdA) expression (A’,C’). Ant, antenna, L1-L3, legs.
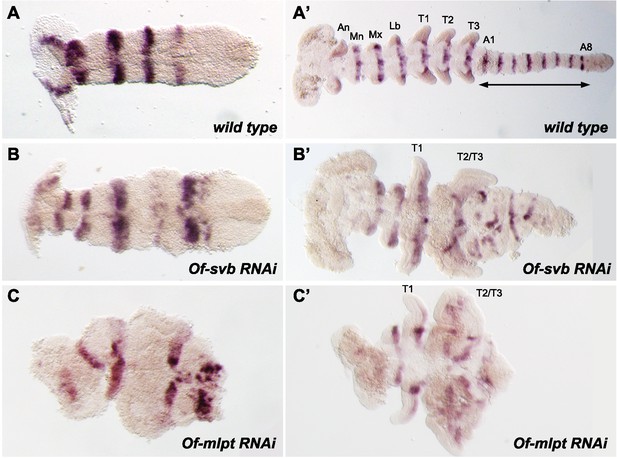
Of-svb and Of-mlpt RNAi leads to defects in embryonic segmentation.
Of-engrailed is expressed in the posterior compartment of each segment in the early (A) and late (B) germband embryos. Knockdown of Of-svb by RNAi results in segment fusion and abnormal segmentation, which is evident in the early germband (A’) and is more pronounced in the late germband (B’) where thoracic segments T2 and T3 are often fused and adjacent abdominal segments appear disordered. The germband is reduced in length, significantly broader in width, and appears incompletely differentiated. Knockdown of Of-mlpt by RNAi also causes disordered patterning, evident in the early germband (C), which is often twisted within the egg shell and exhibits disordered posterior expression. Late germband embryos (C’) exhibit anterior segment loss, frequent T2-T3 fusion, and loss of most abdominal segments. Many embryos appear to lack midline fusion, and exhibit apparent loss of directed embryo elongation, and incomplete differentiation of the germband. An, antennal; Mn, mandibular; Mx, maxillary; Lb, labial segments.

Cuticle defects in hemipteran embryos depleted for svb, mlpt and ubr3.
(A) Detailed view of cuticle and bristles in a wildtype Gerris embryo. (B) Thinning cuticle with missing trichomes in an embryo with reduced Gb-svb. (C) Lateral view of a wild type Oncopeltus hatchling showing characteristic cuticle aspect and pigmentation. Thin, uneven cuticle is observed in knockdown embryos from Of-mlpt RNAi (D), Of-svb RNAi (E) and Of-ubr3 RNAi (F).
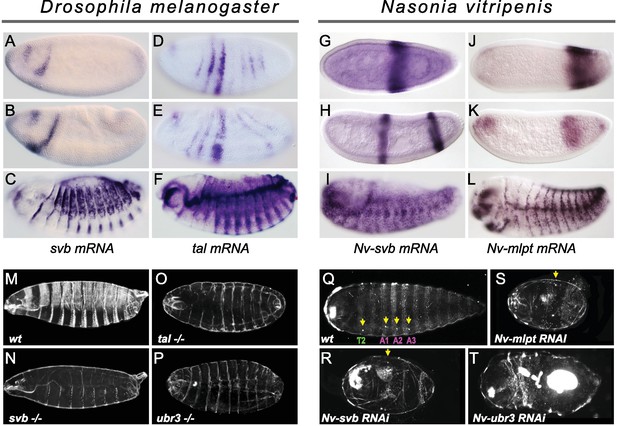
Expression and function of svb, and mlpt/tal in the long germ embryos of Drosophila melanogaster and Nasonia vitripennis.
(A–F) In situ hybridization of Drosophila embryo to svb (A–C) and tal/mlpt (D–F) mRNA. In blastoderm and gastrula embryos, svb mRNA is restricted to two stripes in the head (A,B) while tal is expressed in seven thin stripes in the presumptive abdomen (D,E). At late embryonic stages, svb and tal are expressed in epidermal trichome cells (C,F). (G–L) Expression of Nv-svb (G–I) and Nv-mlpt (J–L) in Nasonia embryo. Nv-svb is expressed in the mid (G) blastoderm in a single broad stripe, and in the late (H) blastoderm in two stripes. Early Nv-mlpt mRNA expression is observed as an anterior cap and a stronger posterior domain (J); anterior expression fades with enrichment of a strong stripe at the posterior as embryogenesis progresses (K). Late Nasonia embryos exhibit widespread Nv-svb and Nv-mlpt expression, with enrichment in a segmental pattern similar to the pattern of trichomes (I, L). (M–P) Cuticles of Drosophila young larvae. (M) Wild type larva showing typical pattern of ventral and dorsal trichomes. Embryos lacking maternal and zygotic tal (O), svb (N), and ubr3 (P) completely lack embryonic trichomes, and exhibit general cuticle defects. (Q–T) Cuticles of Nasonia larvae. (Q) Wild type larva with 4 pairs of spiracles (yellow arrowheads), on thoracic segment T2, and abdominal segments A1, A2 and A3. Cuticles of Nv-mlpt (S) and Nv-svb (R) RNAi larvae are extremely truncated with loss/fusion of most abdominal segments. Fusion of remaining anterior segments are also detected in Nv-mlpt embryos with only one remaining spiracle, Nv-svb larva shows fusion of thoracic segments. Nv-ubr3 RNAi larva exhibit dramatic phenotypes with little or no cuticle. Milder phenotype (T) includes a shortened larva with a thin cuticle decorated with few denticles on the anterior side.

Maternal and zygotic depletion of svb, tal or ubr3 does not affect embryonic segmentation in Drosophila.
Immunostaining for the Wg protein shows the segmentation profile of stage-10 embryos and highlights the correct pattern for tal (B), svb (C) and ubr3 (D) mutant embryos compared to control (A).
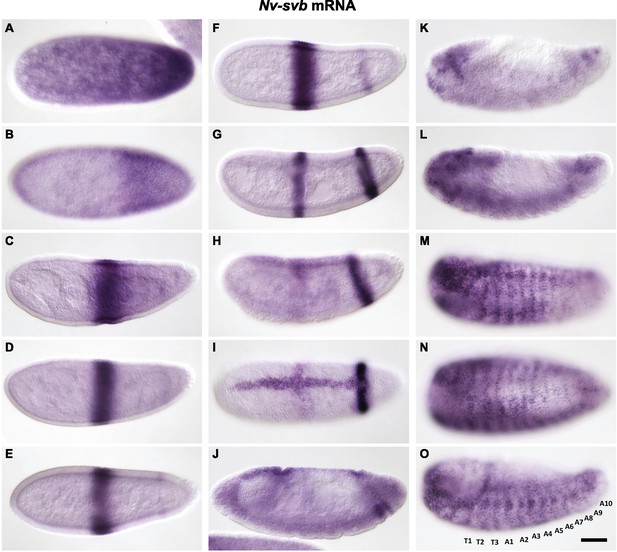
Expression of Nv-svb during embryogenesis.
(A) Maternal Nv-svb is broadly expressed in freshly laid embryos, with obvious posterior enrichment. (B) During blastoderm stages, posterior expression resolves into a broad band, detached from the posterior and clear behind the stripe. (C, D) Broad low-level blastoderm expression clears, leaving strong central Nv-svb expression resembling that of Nv-Kr. (E–F) At cellularization, the central domain is refined and sharpened, and a faint posterior band begins to emerge, at approximately the same position as an Nv-eve posterior stripe, which demarcates the anlage that will give rise to six posterior segments. At gastrulation, a dorsal stripe of Nv-svb emerges as the central stripe of Nv-svb fades and the posterior stripe darkens (H–J) and eventually splits into two discrete stripes (J). At germband retraction, strong staining is evident in the head and in spots along the ventral side of the embryo (K). During dorsal closure, epidermal expression starts as ventral segmental stripes, which extend dorsally and prefigure the pattern of cuticle trichomes; head expression remains strong throughout the remainder of embryogenesis (L–O).
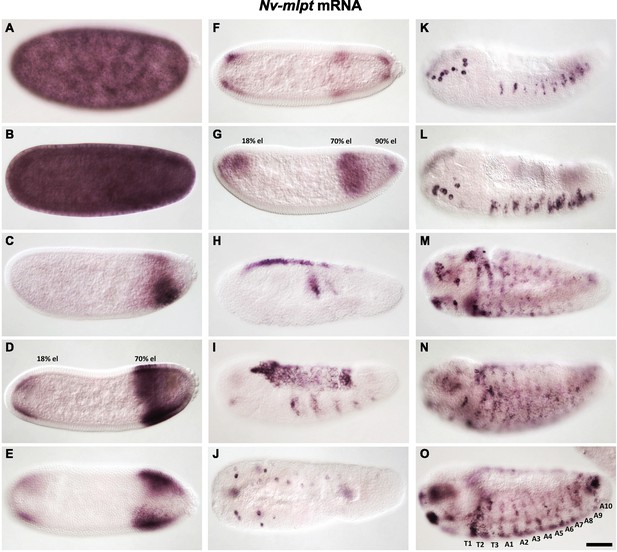
Detailed expression of Nv-mlpt throughout Nasonia embryogenesis.
Maternal expression of Nv-mlpt is strong (A), persisting for several nuclear divisions in syncytium (B). This expression clears approaching cellularization (C), leaving a wedge shaped posterior domain that is slightly withdrawn from the posterior pole. Slightly later, an anterior domain arises, and the stronger posterior domain extends to the posterior end (seen from both lateral (D) and dorsal (E) view). At cellularization, anterior expression remains, while posterior expression has resolved into two distinct stripes: an anterior broad stripe, and a thin posterior stripe at the extreme posterior (F, G). At gastrulation, dorsal expression of Nv-mlpt is apparent, as well as two lateral stripes of expression (H); dorsal expression persists through germband extension, when new ectodermal stripes arise (I). At germband retraction, strong spots of expression are evident in the head and ventrally surface, in nearly every segment (J; dorsal view K). As dorsal closure begins (L), expression is restricted to these spots, while at the end of dorsal closure, expression is seen throughout the embryo, including segmental stripes prefiguring ventral trichome belts, and in strong spots in the head (M–O).
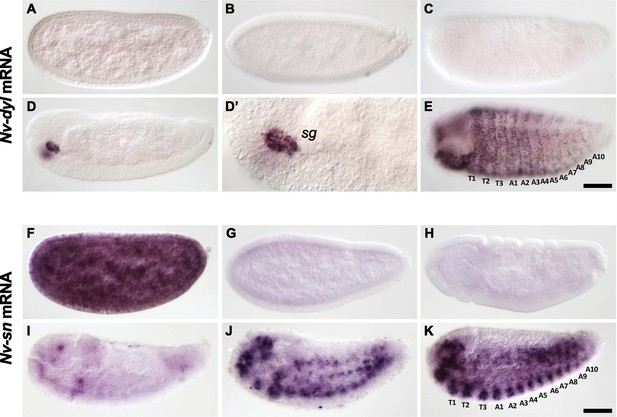
Embryonic expression in Nasonia of dusky-like and singed, two Svb epidermal targets in Drosophila.
(A–E). Embryonic expression of Nv-dusky-like (Nv-dyl) mRNA. Nv-dyl is faintly expressed in freshly laid embryos (A) then disappears (B, C). Nv-dyl is observed again only at dorsal closure with strong expression in discrete structures bilaterally in the head (D,D’) (‘SG’- salivary gland). Later embryos exhibit staining in segmental stripes with additional puncta prefiguring the bristles of the larval cuticle (E). (F–K) Embryonic expression of Nv-singed (Nv-sn). Nv-sn mRNA maternal expression is strong, with ubiquitous staining in freshly laid embryos (F). Signal rapidly fades at cellularization (G) and through gastrulation (H). At germband retraction (I), dots of Nv-sn expression are seen in the head, in the most posterior segment, and along the ventral side, prefiguring segmental dots that darken during dorsal closure (J). Dots correspond to nervous system (ventral) and putative mesodermal derivatives (dorsally). At the end of embryogenesis (K), segmental stripes are seen along the ventral epidermis with strong staining in the head.
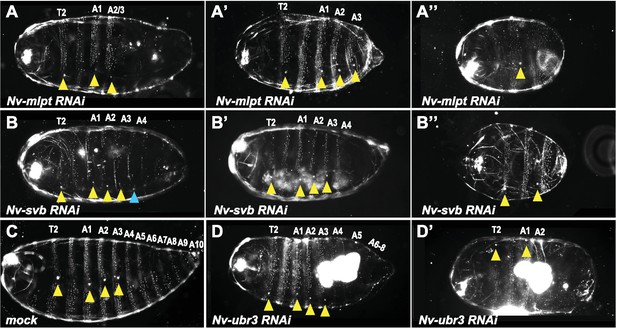
Phenotypes of increasing strength for Nv-mlpt-RNAi, Nv-svb-RNAi, Nv-ubr3-RNAi in Nasonia embryos.
(A–A”) Cuticle phenotypes of Nv-mlpt RNAi embryos. Mild RNAi phenotypes from Nv-mlpt knockdown result in loss of posterior abdominal segments, which can be seen as reduction in larval cuticle length and loss of posterior trichomes (A). Moderate RNAi phenotypes cause more severe posterior truncation and segment fusions (A’). Severe RNAi phenotypes from Nv-mlpt RNAi (A”) cause loss of most posterior segments, fusion of remaining abdominal segments, and anterior fusions in thoracic segments. (B–B”) Cuticle phenotypes of Nv-svb RNAi embryos. (B) Mild RNAi phenotypes from Nv-svb RNAi result in shortened larval cuticles with missing posterior trichomes; occasionally, ectopic spiracles (normally restricted to T2 and A1-A3) are apparent on A4 (blue arrowhead). Moderate phenotypes (B’) are even more truncated, with loss of posterior structures. Severe phenotypes (B”) to dramatically truncated embryos, with loss of all posterior abdominal segments, fusions of remaining anterior abdominal segments, but normal anterior terminal structures. Yellow arrowheads indicate remaining T2 spiracle and single remaining spiracle associated with likely fused A1-A3 abdominal segments. Nv-ubr3 RNAi resulted in severe defects in cuticle secretion, and very fragile embryos which were impossible to recover intact. Rare mildest phenotypes are shown (C’–C”) compared to mock RNAi (C) for normal morphology reference. Wild type and mock RNAi larval cuticles possess four spiracles on segments T2, A1, A2 and A3 (yellow arrowheads), and stereotypic bands of trichomes on thoracic and abdominal segments. Mild RNAi phenotype of Nv-ubr3 knockdown causes posterior truncation of the embryo, and loss of posterior trichomes (posterior to A3). Anterior terminal structures appear intact.
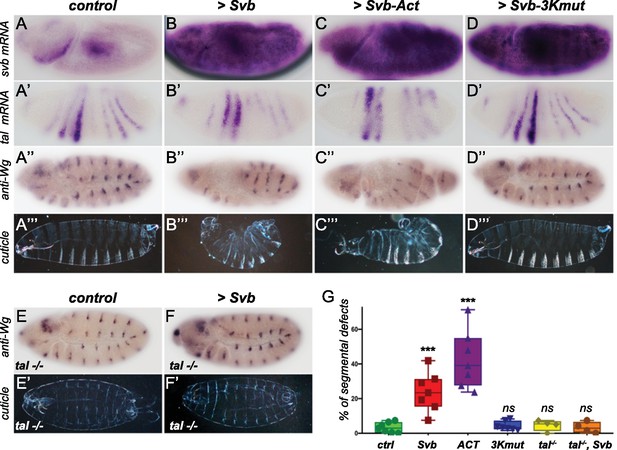
Reawakening svb expression in the early Drosophila embryo affects segmentation.
Top panels show in situ hybridization of svb (A–D) and tal (A’–D’) mRNA and anti-Wingless (Wg) immunostaining (A”–D”) at gastrulation stage in control conditions (nullo >GFP) and following the ectopic expression (driven by nullo-Gal4) of wild type Svb (B–B”), Svb-ACT (C–C”) and Svb-3Kmut (D–D”), which mimics or prevents Pri/Ubr3-mediated processing of Svb, respectively. (A’’’–D’’’) show cuticle preparations of control (A’’’), nullo >Svb (B’’’), nullo >Svb ACT (C’’’) and nullo >Svb-3Kmut (D’’’) embryos. (E–F’) panels show immunostaining for the Wingless protein and cuticle preparations of control (E–E’) and svb ectopic expression (nullo >Svb) (F–F’) in a tal null genetic background. tal mutant embryos display characteristic trichome loss and cuticle defects. (G) Quantification of segmental defects for each genotype. Data were analyzed by one-way ANOVA. ***, p-value<0,002; ns, non-significant. Total numbers of embryos are 177 (ctrl), 62 (Svb), 621(Act), 413 (3Kmut), 223 (tal-/-) and 138 (tal-/-, Svb). Source data for Figure 6G are found in Source Data File 1.
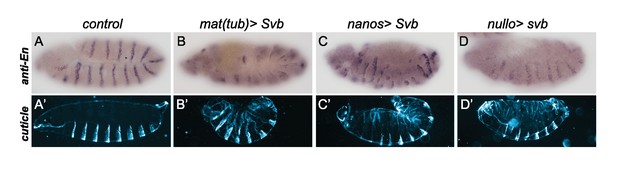
Early ectopic expression of svb using different drivers triggers segmentation defects.
Expression of svb from oogenesis using mat-Gal4 (B,B’) and nanos-Gal4 (C–C’), or from blastoderm stage using nullo-Gal4 (D,D’), alters segmentation as seen on embryos stained with anti-Engrailed (En) antibody (A–D) and by cuticle analysis (A’–D’).
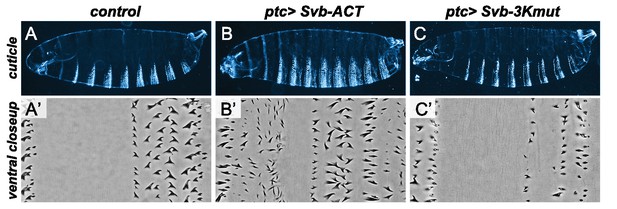
Effect of modified forms of the Svb protein on epidermal trichome formation.
UAS-GFP (control) (A,A’), UAS-Svb-ACT (B,B’) and UAS-Svb-3Kmut (C,C’) were expressed in the embryonic epidermis under the control of the ptc-Gal4 driver. Top rows show whole embryo cuticles (A–C), the bottom row shows close-ups in the ventral region of the third abdominal segment (A’–C’). Svb-ACT, which lacks the N-terminal repressor domain and thus mimics the processed form of Svb, acts as a constitutive activator of transcription and triggers the production of ectopic trichomes. In contrast, Svb-3Kmut -bearing mutations on the 3 Lysines ubiquitinated by Ubr3 in response to Tal peptides- behaves as a repressor and counteracts endogenous Svb activity, resulting in loss of trichomes.
Tables
Summary of Tribolium phenotypes resulting from RNAi-mediated depletion of mlpt, Tc-Ubr3, Tc-Svb, as well as those observed in Tc-Svb CRISPR mutants.
In each case, a total of 20 animals were scored. Data show the average number of deleted abdominal segments, missing terminal appendages (urogomphi) and number of pairs of extra legs. Cuticle defects were scored as normal-looking (-), mild (+) and strong (+++) thinning. For leg length, the distance from coxa/trochanter joint to leg tip (see Figure 1) was measured in segment T3.
| Deleted abdominal segments | Urogomphi missing | Thoracic leg length (µm) | Extra legs | Cuticle thinning | |
|---|---|---|---|---|---|
| Wild type | 0 | 0 | 183 | 0 | - |
| mlpt-RNAi | 3.8 | 2 | 170 | 4.3 | - |
| Tc-ubr3 RNAI | 5.1 | 2 | 112 | 3.9 | + |
| Tc-svb RNAi | 0.5 | 1.5 | 102 | 3.2 | + |
| Tc-svb CRISPR | 1.0 | 1.7 | 122 | 1.65 | +++ |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | ovo/svb | NA | FLYB:FBgn0003028 | |
| Gene (Drosophila melanogaster) | tal | NA | FLYB:FBgn0087003 | |
| Gene (Drosophila melanogaster) | Ubr3 | NA | FLYB:FBgn0260970 | |
| Gene (Tribolium castaneum) | Tc-svb | this paper | Genbank: MG913606 | |
| Gene (Tribolium castaneum) | mlpt | NA | GenBank: AM269505.1 | |
| Gene (Tribolium castaneum) | Tc-Ubr3 | NA | NCBI Ref Seq: XM_964327 | beetlebase: TC005949 |
| Gene (Oncopeltus fasciatus) | Of-svb | this paper | GenBank: MH181832 | |
| Gene (Oncopeltus fasciatus) | Of-mlpt | this paper | GenBank: MH181830 | |
| Gene (Oncopeltus fasciatus) | Of-Ubr3 | this paper | GenBank: MH181827 | |
| Gene (Gerris buenoi) | Gb-svb | this paper | GenBank: MH011417 | |
| Gene (Gerris buenoi) | Gb-mlpt | this paper | GenBank: MH699965 | |
| Gene (Gerris buenoi) | Gb-Ubr3 | this paper | GenBank: MH011418 | |
| Gene (Nasonia vitripennis) | Nv-svb | this paper | GenBank: MH181831 | |
| Gene (Nasonia vitripennis) | Nv-mlpt | this paper | GenBank: MH181829 | |
| Gene (Nasonia vitripennis) | Nv-Ubr3 | this paper | GenBank: MH181828 | |
| Strain, strain background (Nasonia vitripennis) | AsymCx | PMID: 20075255 | ||
| Genetic reagent (D. melanogaster) | FM7C, Kr > GFP | Bloomington Drosophila Stock Center | BDSC: 5193; FLYB: FBst0005193; RRID:BDSC_5193 | FlyBase symbol: Df(1)JA27/FM7c, P{w[+mC]=GAL4 Kr.C}DC1, P{w[+mC]=UAS GFP.S65T}DC5, sn[+] |
| Genetic reagent (D. melanogaster) | TM6B, ubi-GFP | Bloomington Drosophila Stock Center | BDSC: 4887; FLYB: FBst0004887; RRID:BDSC_4887 | FlyBase symbol: w[1118]; Df(3L)Ly, sens[Ly-1]/TM6B, P{w[+mW.hs]=Ubi GFP.S65T}PAD2, Tb[1] |
| Genetic reagent (D. melanogaster) | TM3, twist-GAL4 > GFP | Bloomington Drosophila Stock Center | BDSC: 6663; FLYB: FBst0006663; RRID:BDSC_6663 | FlyBase symbol: w[1118]; Dr[Mio]/TM3, P{w[+mC]=GAL4 twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb[1] Ser[1] |
| Genetic reagent (D. melanogaster) | nullo-GAL4 | Bloomington Drosophila Stock Center | BDSC:26875; FLYB:FBtp0018484; RRID:BDSC_26875 | FlyBase symbol: P{nullo-GAL4.G}5.20 |
| Genetic reagent (D. melanogaster) | nos-GAL4 | Bloomington Drosophila Stock Center | BDSC:4937; FLYB:FBtp0001325; RRID:BDSC_4937 | FlyBase symbol: P{GAL4::VP16- nos.UTR}CG6325MVD1 |
| Genetic reagent (D. melanogaster) | ptc-GAL4 | Bloomington Drosophila Stock Center | BDSC:2017; FLYB:FBti0002124; RRID:BDSC_2017 | FlyBase symbol: P{GawB}ptc559.1 |
| Genetic reagent (D. melanogaster) | pri[1] | PMID:17486114 | FLYB:FBal0198099 | Flybase symbol: talS18 |
| Genetic reagent (D. melanogaster) | tal[S18.1] | PMID:17486114 | FLYB:FBal0241050 | Flybase symbol: talpri-1 |
| Genetic reagent (D. melanogaster) | pri[4] | gift from Y. Kageyama | ||
| Genetic reagent (D. melanogaster) | pri[5] | gift from Y. Kageyama | ||
| Genetic reagent (D. melanogaster) | svb[R9] | PIID: 12915226 | FLYB:FBal0151651 | Flybase symbol: ovo[svb-R9] |
| Genetic reagent (D. melanogaster) | ovo[D1] | PMID: 17246182 | BDSC: 23880; FLYB: FBst0023880; RRID:BDSC_23880 | Flybase symbol: ovo[D1] |
| Genetic reagent (D. melanogaster) | svb[PL107] | PMID: 11744370 | DGGR:106675; FLYB: FBst0305341; RRID:DGGR_106675 | Flybase symbol: ovo[PL107] |
| Genetic reagent (D. melanogaster) | Ubr3B | PMID: 26383956 | FLYB:FBal0013375 | Flybase symbol: Ubr3[B] |
| Genetic reagent (D. melanogaster) | UAS-GFP | Bloomington Drosophila Stock Center | FLYB:FBal0129171 | FlyBase symbol: w[*]; P{w[+mC]=UAS GFP .S65T}Myo31DF[T2] |
| Genetic reagent (D. melanogaster) | UAS-svb::GFP | PMID: 20647469 | FLYB: FBal0319860 | FlyBase symbol: ovoUAS.svb.GFP |
| Genetic reagent (D. melanogaster) | UAS-pri | PMID: 17486114 | BDSC: 1521; FLYB:FBti0003040; RRID:BDSC_1521 | FlyBase symbol: talUAS.cKa |
| Genetic reagent (D. melanogaster) | UAS-svbACT::GFP | this paper | FLYB:FBal0248431 | |
| Genetic reagent (D. melanogaster) | UAS-svb-3Kmut::GFP | this paper | FLYB:FBal0241056 | |
| Antibody | anti-Wingless | Developmental Studies Hybridoma Bank | (1:100) | |
| Antibody | anti-Ubx-AbdA | Developmental Studies Hybridoma Bank | (1:5) | |
| Antibody | anti-Dll abbit polyclonal | DSHB Cat# 4d4; RRID:AB_528512 | (1:200) r | |
| Antibody | anti-Dig AP Fap (polyclonal sheep) | Roche | DSHB Cat# UBX/ABD-A FP6.87; RRID:AB_10660834 | (1:2000) |
| Antibody | anti-mouse-HRP (rabbit polyclonal) | Promega | (1:1000) | |
| Antibody | anti-rabbit-HRP (donkey polyclonal) | Jackson Immuno Research | Roche Cat# 11093274910; RRID:AB_514497 | (1:500) |
| Antibody | anti-mouse biotinylated (goat polyclonal) | Vector Laboratories | Promega Cat# W4011; RRID:AB_430833 | (1:500) |
| Recombinant DNA reagent | pUASp-Svb::GFP (plasmid) | PMID:17486114 | Jackson ImmunoResearch Labs Cat# 711-035-152; RRID:AB_10015282 | |
| Recombinant DNA reagent | pUASp-SvbAct::GFP (plasmid) | this paper | Vector Laboratories Cat# BA-9200; RRID:AB_2336171 | Progenitors: PCR, pUASp-Svb::GFP |
| Recombinant DNA reagent | pUASp-Svb-3Kmut::GFP (plasmid) | this paper | Progenitors: pAc-SvbK7; pUASp-Svb::GFP | |
| Recombinant DNA reagent | pCR-Topo (plasmid) | Qiagen | ||
| Recombinant DNA reagent | pBluescript (plasmid) | Stratagene | ||
| Recombinant DNA reagent | pGEM-Teasy (plasmid) | Promega | Quiagen Cat#: 231122 | |
| Recombinant DNA reagent | pBac (3xP3-EGFPafm) (plasmid) | gift from E. Wimmer | Stratagene Cat#: 212205 | Flybase symbol: PBac{3xP3-EGFPafm} |
| Recombinant DNA reagent | pBME(TcU6b-BsaI) (plasmid) | gift from A. Giles | Promega Cat#: A1360 | Original gRNA expression vector with Bsa1 sites |
| Recombinant DNA reagent | pSLfa(Hsp-p-nls-Cas9-3’UTR)fa (plasmid) | gift from A. Giles | FLYB: FBtp0014061 | Cas9 expression vector |
| Recombinant DNA reagent | Tc-U6b-sim ZS1 (plasmid) | Rode and Klingler, unpublished | sim gRNA expression vector | |
| Sequence-based reagent | seeSupplementary file 1B for a complete list of oligonucleotides used in this paper | |||
| Commercial assay or kit | DIG RNA Labeling kit | Roche | ||
| Commercial assay or kit | NBT-BCIP solution | Roche | ||
| Commercial assay or kit | In-Fusion HD Cloning Kit | Clontech | Roche Cat#: 11 277 073 910 | |
| Commercial assay or kit | MEGAscript RNA kit | ThermoFischer | Sigma Cat#: 72091 | |
| Chemical compound, drug | Blocking reagent | Roche | Takara Cat#: 21416 | |
| Chemical compound, drug | 3,3′-Diaminobenzidine tetrahydrochloride hydrate | Sigma | ThermoFischer Cat#: AM1626 | |
| Software, algorithm | Next-RNAi | http://www.nextrnai.org | Roche Cat#: 11 096 176 001 | |
| Software, algorithm | Primer3 | https://primer3plus.com | Sigma Cat#:32750 | |
| Software, algorithm | MacVector | https://macvector.com | ||
| Software, algorithm | Prism 8 | https://www.graphpad.com/ | Primer3Plus; RRID:SCR_003081 | |
| Software, algorithm | Photoshop CC 2019 | https://www.adobe.com/ | MacVector; RRID:SCR_015700 | |
| Software, algorithm | Illustrator CC 2019 | https://www.adobe.com/ | GraphPad Prism; RRID:SCR_002798 | |
| Software, algorithm | Acrobat Pro DC | https://www.adobe.com/ | Adobe Photoshop; RRID:SCR_014199 | |
| Software, algorithm | Axiovision 4.6.3.SP1 | Zeiss | Adobe Illustrator; RRID:SCR_010279 |
Additional files
-
Source data 1
Source data for charts in Figures 1G and 6G, and Figure 4—figure supplement 1–3
- https://doi.org/10.7554/eLife.39748.031
-
Supplementary file 1
Supplementary information.
(A) Genes identified in the genome wide iBeetle RNAi screen that had phenotypes resembling those of mlpt. iB 00966 and 09278, had the most highly penetrant RNAi phenotype with the strongest resemblance to those of mlpt. NCBI annotates these as belonging to a single locus LOC657900 encoding a 6592 bp mRNA (accession number XM_964327) that corresponds to Tc-ubr3. (B) Oligonucleotides used in the manuscript. Nucleotides shown in red indicate tags of parts of T7 (3’ primer) and SP6 (5’ primer) promoter sequences attached to gene-specific sequences for nested PCR. Nucleotides shown in blue highlight minimal T7 promoter used in subsequent in vitro T7 RNA polymerase transcription. (C) Oligos used for generating Tc-svb CRISPR mutant. (D) Plasmids used for generating Tc-svb CRISPR mutant.
- https://doi.org/10.7554/eLife.39748.032
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39748.033





