Spliceosome factors target timeless (tim) mRNA to control clock protein accumulation and circadian behavior in Drosophila
Figures
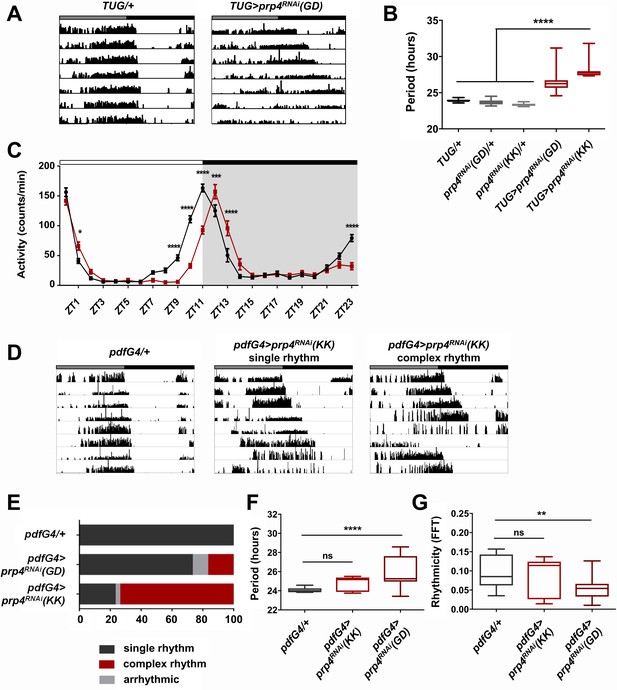
Pre-mRNA splicing factor four is a new regulator of the circadian clock.
(A) Representative activity records of free-running fly behavior upon downregulation of prp4 in tim+ neurons. Dicer2 (Dcr2) was co-expressed with the RNAi transgenes to increase the knockdown efficiency. Genotypes are indicated on top of each panel. The gray and black bars indicate the subjective day and night, respectively. (B) The lengthening of free-running circadian period is significant for two independent prp4 RNAi lines (GD and KK). ****p ≤ 0.0001 relative to heterozygous controls by one-way ANOVA and Tukey’s post hoc test, n = 8–24. (C) Downregulation of prp4 in tim+ cells affects morning and evening anticipation in 12 hr:12 hr light:dark (LD) conditions. The activity profile of control (TUG; Dcr2/+) flies is in black, while the activity profile of experimental (TUG; Dcr2 >prp4RNAi(GD)) flies is in red. The white and black bars indicate light and dark conditions, respectively. *p ≤ 0.05, ***p ≤ 0.001,****p ≤ 0.0001 to control (TUG; Dcr2/+) for each ZT range by two-way ANOVA and Sidak’s post hoc test. Data represent mean ±SEM (n = 31–32). (D) Activity records demonstrate 7 days of representative free-running rhythms of prp4 knockdown flies. Dicer2 (Dcr2) was co-expressed with the RNAi transgenes to increase the knockdown efficiency. Genotypes are indicated on top of each panel. The gray and black bars indicate the subjective day and night, respectively. (E) Knockdown of prp4 in LNv pacemaker cells causes complex behavioral periods (p < 0.0001 by χ2 analysis, n = 19–38). (F, G) Knockdown of prp4 in LNvs lengthens circadian period and decreases rhythm strength. n.s., not significant at the 0.05 level, **p ≤ 0.01, ****p ≤ 0.0001 to control (pdfG4; Dcr2/+) by one-way ANOVA and Holm-Sidak’s post hoc test. Only rhythmic flies (FFT > 0.01) were analyzed (n = 9–22). In panels (B), (F) and (G), the boxes extend from the 25th to 75th percentiles, the line within the box is plotted at the median and whiskers extend from the lowest to highest value.
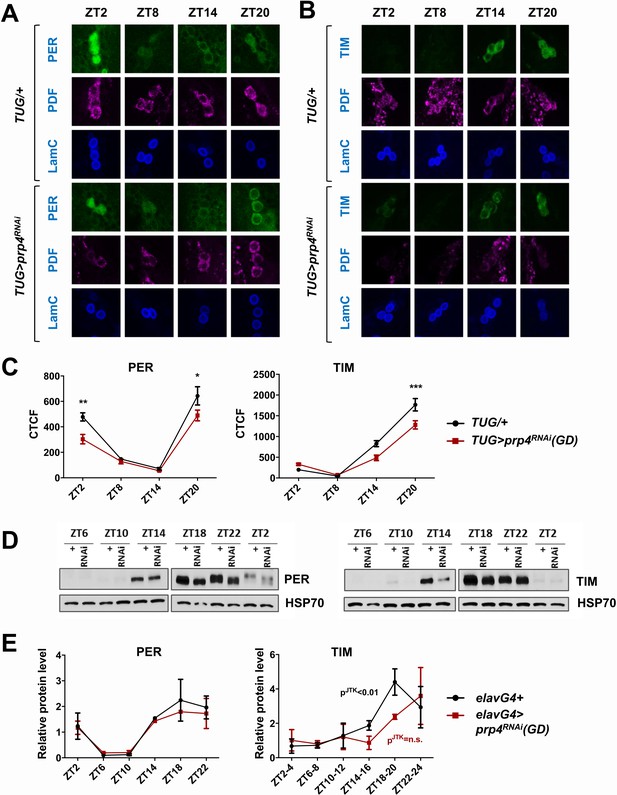
PRP4 is required for robust TIM and PER cycling.
(A, B) Cycling of PER and TIM is disrupted in s-LNvs of prp4 knockdown flies. Adult brains were dissected at time points indicated and immunostained with PER or TIM (green), PDF (magenta) and LaminC (blue) antibodies. Dicer2 (Dcr2) was co-expressed with the prp4 RNAi transgene to increase its knockdown efficiency. Genotypes are indicated on the sides of each panel. The displayed images are representative of 2–3 independent experiments. (C) Corrected Total Cell Fluorescence (CTCF) was used to quantitatively assess the change in levels of PER and TIM in s-LNvs. The signal from both the nucleus and the cytoplasm was used to calculate CTCF. 10–21 cells from 5 to 8 brains were analyzed for ZT2, ZT14 and ZT20 and 8–10 cells from 3 to 5 brains for ZT8. Images were taken with identical confocal settings. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 to control (TUG;Dcr2/+) for each ZT time point as determined by two-way ANOVA and Sidak’s post hoc test. Error bars = ± SEM. (D) PER phosphorylation is decreased and cycling of TIM is blunted in fly heads with pan-neuronal knockdown of prp4. Dicer2 (Dcr2) was co-expressed with the prp4 RNAi transgene to increase its knockdown efficiency. Adult fly heads were collected at indicated zeitgeber (ZT) time points in a 12 hr:12 hr light:dark cycle. Representative western blots probed for PER (right) or TIM (left) are shown. HSP70 was used as a loading control. (E) Total PER and TIM levels were quantified from western blots in (D). JTK cycle analysis identified significant cycling (pJTK ≤ 0.01) for control (elavGal4; Dcr2/+) and not significant (n.s at the 0.05 level) cycling for samples with pan-neuronal prp4 knockdown. Data represent mean ±SEM (n = 2–3 independent experiments).
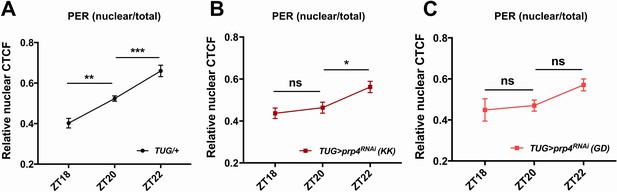
prp4 downregulation delays night-time accumulation of PER in nuclei of s-LNvs.
The ratio of nuclear to total PER levels indicates reduced nuclear accumulation of PER at night upon prp4 knockdown with 2 RNAi lines, KK (B) and GD (C), relative to control (A). Corrected Total Cell Fluorescence (CTCF) was used to quantify both the total (cytoplasm +nucleus) and the nuclear PER levels in s-LNvs at indicated ZT time points. 15–17 cells (A) 14–17 cells (B) or 8–11 cells (C) from at least four brains were used for quantification. Dicer2 (Dcr2) was co-expressed with the prp4 RNAi transgene to increase its knockdown efficiency. Images were taken with identical confocal settings. n.s., not significant at the 0.05 level, *p ≤ 0.05, p** ≤ 0.01, p*** ≤ 0.001 to control (TUG;Dcr2/+) as determined by two-way ANOVA and Sidak’s post hoc test. Error bars = ±SEM.
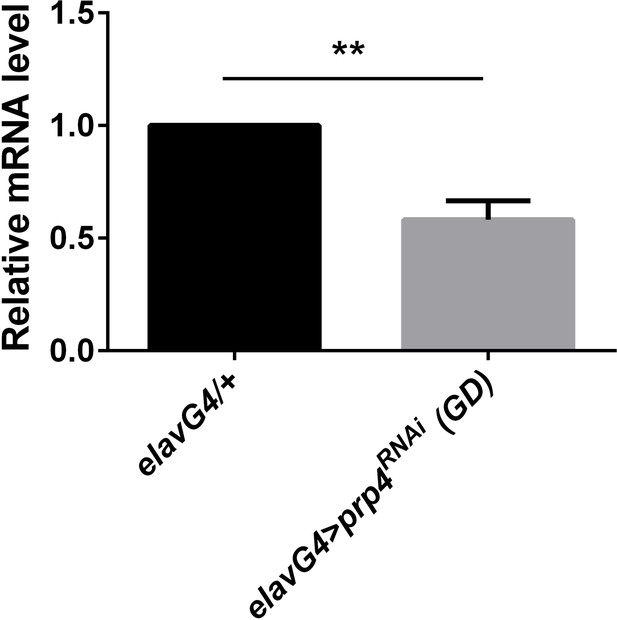
prp4 is efficiently knocked down with the GD RNAi line.
Pan-neuronal knockdown of prp4 decreases mRNA levels of prp4 as calculated with Student’s t test, p** ≤ 0.01. Dicer2 (Dcr2) was co-expressed with the prp4 RNAi transgene to increase its knockdown efficiency. Three independent qPCR experiments were performed in triplicate, normalized to rp49 and analyzed using the ∆∆Ct method. Error bars = SEM.
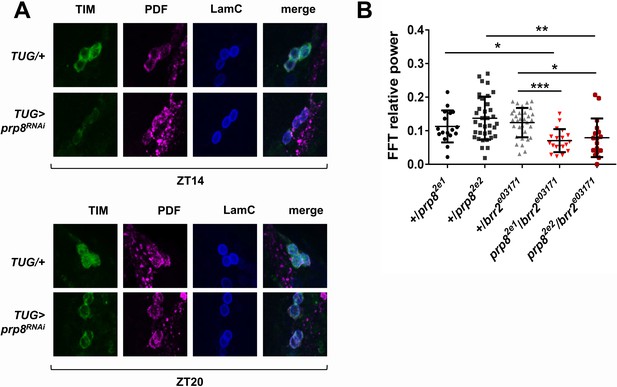
PRP8 regulates TIM levels and the strength of rest:activity rhythms.
(A) TIM levels are decreased in s-LNvs of prp8 knockdown flies. Adult brains were dissected at ZT14 and ZT20 on the 4th day in LD cycle and immunostained with TIM (green), PDF (magenta) and LaminC (blue) antibodies. Genotypes are indicated on the sides of each panel. Displayed images are representative of two independent experiments. (C) Trans-heterozygous prp8/brr2 mutants have weaker circadian rhythms compared to their heterozygous controls, p* ≤ 0.05, p** ≤ 0.01, p*** ≤ 0.001 by one-way ANOVA, Tukey post hoc test. All FFT value were used in the analysis, including the arrhythmic ones (FFT < 0.01). Error bars represent mean ±SEM (n = 17–36). Figure 3 is related to Table 1.
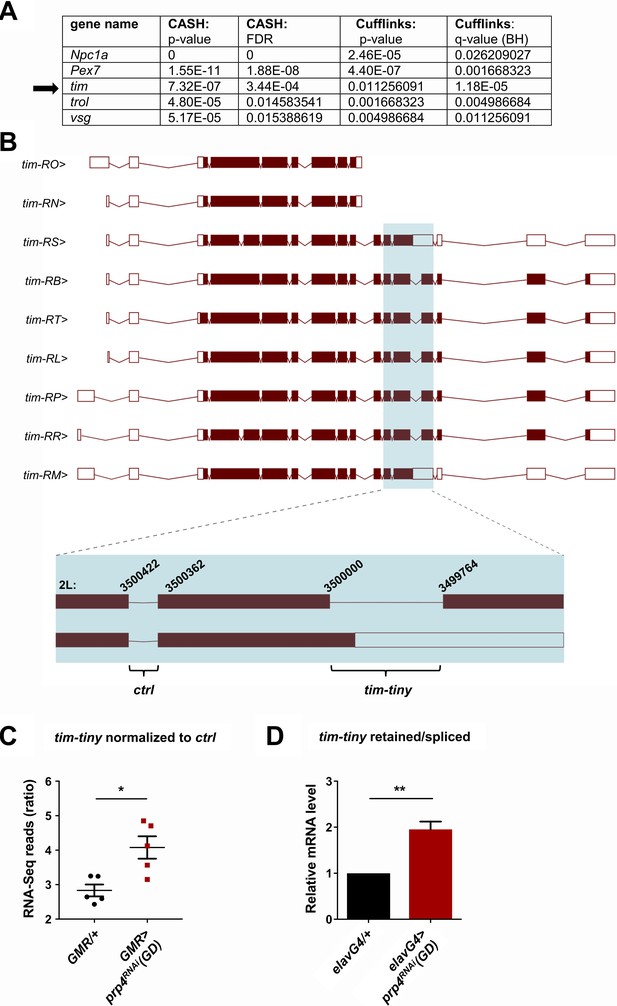
PRP4 regulates tim splicing.
(A) Only five genes were identified as differentially spliced upon prp4 downregulation with both CASH and Cufflinks/differential psi (percent spliced in) pipelines. For each gene, the corresponding p-value and False Discovery Rate (FDR) or q-value as determined by Benjamini-Hochberg (BH) procedure are reported. (B) tim isoforms (image adapted from Ensembl Fruitfly release 92, genome assembly BDGP6) are displayed. The boxes indicate exons, with filled boxes (brown) representing protein-coding sequences. The region of interest is enlarged (blue box) and depicts a constitutively spliced intron (‘ctrl’) and the intron that gets retained upon prp4 knockdown (‘tim-tiny’). The chromosomal coordinates of these introns are indicated at their respective exon-intron junctions. (C) tim-tiny retention was revealed by RNA-Seq analysis in samples with prp4 downregulated (GMR > prp4 RNAi). The number of RNA-Seq reads across the tim-tiny intron normalized to the number of reads across the ctrl intron is higher in prp4 knockdown flies (GMR > prp4RNAi) compared to controls (GMR/+). Data represent five independent biological replicates. Error bars represent mean RNAiSEM. *p ≤ 0.0001 as determined by CASH (refer to panel A). (D) An increase in intron retention in flies with pan-neuronal prp4 knockdown was confirmed with qPCR analysis. Dicer2 (Dcr2) was co-expressed with the prp4 RNAi transgene to increase its knockdown efficiency. Data represent four independent biological replicates, with technical triplicates performed during the qPCR step for each replicate. **p ≤ 0.01 to control (elavGal4; Dcr2/+) as determined by Student’s t test. Data represent mean ±SEM.
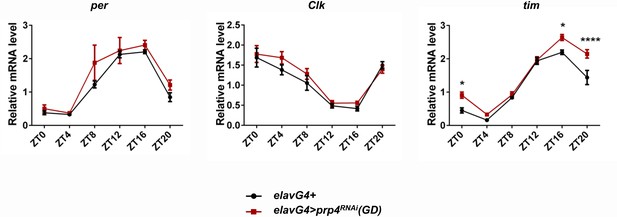
Pan-neuronal knockdown of prp4 increases tim expression.
per and Clk expression is unchanged while tim expression is increased at night (ZT16-ZT0) in the heads of flies with pan-neuronal prp4 downregulation (elavGal4; Dcr2 > prp4RNAi(GD)) as compared to control (elavGal4; Dcr2/+); p* ≤ 0.05, p**** ≤ 0.001 by by two-way ANOVA and Sidak’s post hoc test.
Three independent qPCR experiments were performed in triplicates, normalized to rp49 and analyzed using the ∆∆Ct method. Error bars = SEM.
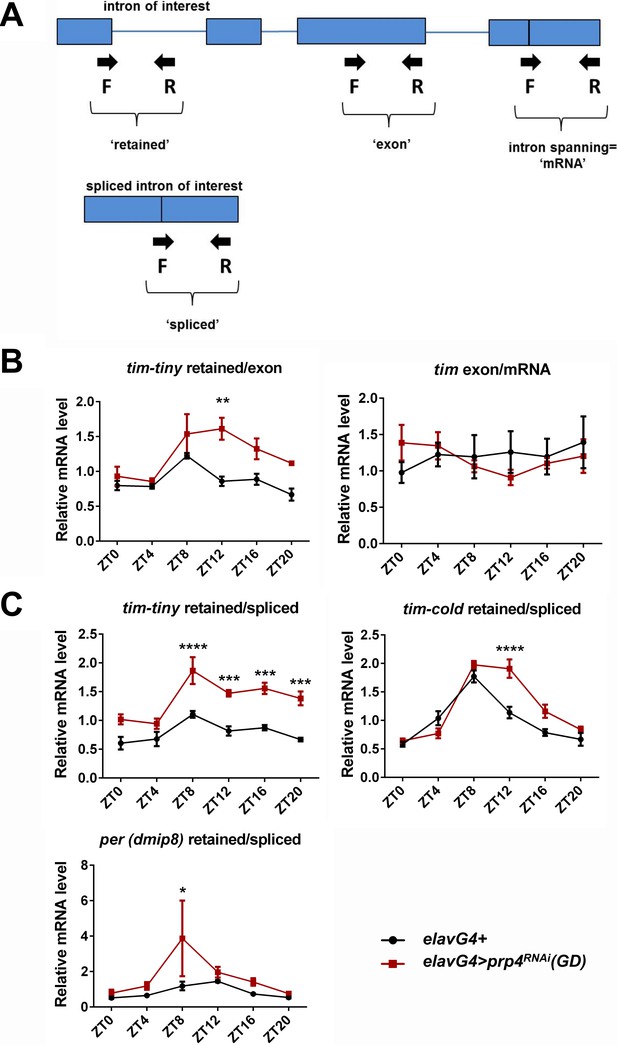
Analysis of the splicing of tim-tiny, tim-cold and per introns.
(A) Schematic for qPCR-based analysis of tim-tiny splicing. Arrows indicate forward and reverse primers. Exons are depicted as boxes and introns are shown as lines. (B) On the left, normalization of the tim-tiny ‘retained’ signal to ‘exon’ primer signal verifies the increased tim-tiny retention in elavGal4; Dcr2 > prp4 RNAi (GD) flies (red) compared to elavGal4; Dcr2/+ flies (black). On the right, no difference is detected between the non-exon-spanning (‘exon’) and exon-spanning (‘total’) tim primer sets. Data represent 3–4 biological replicates, with technical triplicates performed during the qPCR step for each biological replicate. p** ≤ 0.01 by two-way ANOVA and Sidak’s post hoc test. (C) Diurnal profiles of tim-tiny, tim-cold and per intron retention in control (elavGal4; Dcr2/+) and prp4 loss-of-function (elavGal4; Dcr2 > prp4 RNAi (GD)) flies were assayed with qPCR and expressed as a ratio of retained to spliced introns at indicated ZT time points. Data represent 3–4 biological replicates, with technical triplicates performed during the qPCR step for each biological replicate. p* ≤ 0.05, p*** ≤ 0.001, p**** ≤ 0.001 by two-way ANOVA and Sidak’s post hoc test.
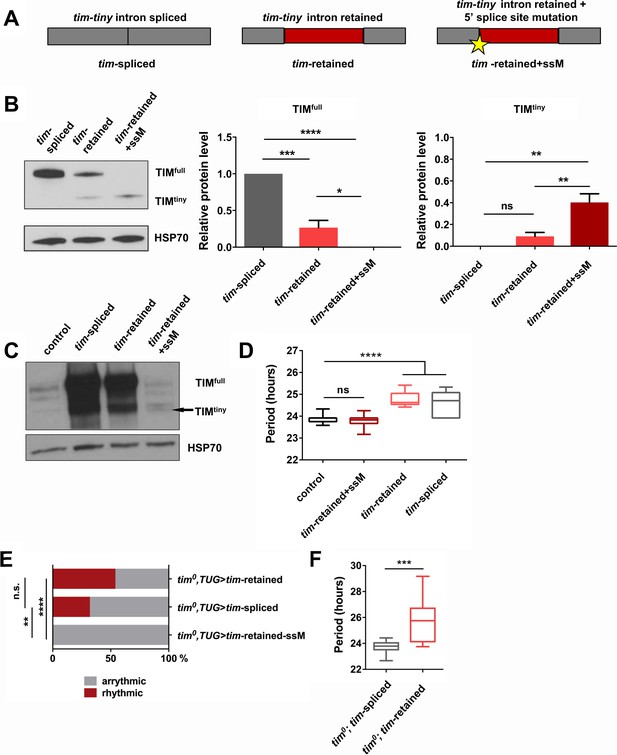
Intron retention in tim decreases TIM levels and affects circadian behavior.
(A) Schematic depiction of three tim cDNA constructs used to assess the effect of tim-tiny retention (red block) on TIM levels. (B) Retention of tim-tiny intron decreases full-length TIM and leads to production of a minor TIMtiny isoform. S2 cells were transfected with constructs described in (A) and western blots of cell lysates were probed with TIM antibody. Western blots are representative of 3 independent experiments. In the panels on the left, total levels of TIM isoforms upon expression of splice-specific cDNA constructs were quantified. TIM levels were normalized to HSP70 and expressed relative to the TIMfull levels in cells overexpressing a fully spliced (tim-spliced) construct. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 by one-way ANOVA and Holm-Sidak’s post hoc test. Data represent mean ±SEM (n = 3). (C) Western blots of head lysates of flies overexpressing tim cDNA with a 5’ splice site mutation in tim-tiny (TUG; Dcr2 > tim-retained+ssM) reveal production of TIMtiny (arrow) and decrease in TIMfull compared to flies overexpressing intronless tim cDNA (TUG; Dcr2 > timspliced) or tim cDNA that includes tim-tiny ((TUG; Dcr2 > tim retained). All flies were collected at ZT10, when endogenous TIM levels are low in control flies (TUG; Dcr2/+). Western blots are representative of 4 independent experiments. (D) Flies overexpressing tim cDNA constructs with 5’ splice site mutation (TUG; Dcr2 >tim-retained+ssM) do not lengthen circadian period. n = 6–26; n.s., not significant at the 0.05 level; ****p ≤ 0.0001 to control (TUG; Dcr2/+) by one-way ANOVA and Tukey’s post hoc test. (E) TUG-driven expression of tim cDNA, with both ‘tim-spliced’ and ‘tim-retained’ constructs, rescues circadian rhythms in tim0 flies. n.s., not significant at the 0.05 level, **p ≤ 0.01, ****p ≤ 0.0001 by pairwise Fischer’s exact test (n = 28–41). (F) TUG-driven rescue of tim0 circadian rhythms with tim cDNA lacking tim-tiny (tim0,TUG > tim spliced) results in shorter periods than with tim cDNA that includes tim-tiny (tim0,TUG > tim retained). ***p ≤ 0.001 by Student’s t test, n = 10–22.
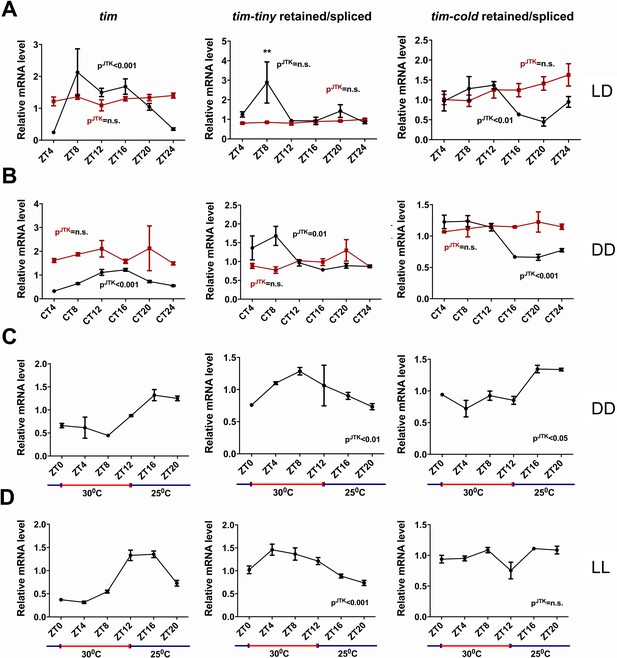
tim splicing is regulated by the clock and by temperature.
Flies were entrained for at least 3 days in 12 hr:12 hr light:dark (LD) conditions and collected in LD (A) or on the first day of transfer to constant darkness (B) at indicated ZT or CT time points, respectively. Splicing of tim-tiny and tim-cold introns was quantified as a ratio of retained to spliced levels using qPCR analysis. Three independent qPCR experiments were performed in triplicate, normalized to rp49 and analyzed using the ∆∆Ct method. pJTK indicates cycling as assessed by JTK cycle analysis for wild-type iso31 flies (black) and per01 circadian mutants (red). tim-tiny intron retention is increased in LD at ZT8 (A) as calculated by two-way ANOVA and Sidak’s post hoc test, p** ≤ 0.01. Wild-type iso31 flies were collected at indicated ZT time points after at least four full days of entrainment in 12 hr:12 hr 30C:25C temperature cycles in constant dark (C) or constant light (D) conditions. Splicing of tim-tiny and tim-cold introns was quantified as a ratio of retained to spliced levels using qPCR analysis. Three independent qPCR experiments were performed in triplicate, normalized to rp49 and analyzed using the ∆∆Ct method. pJTK indicates cycling as assayed by JTK cycle analysis. Error bars = SEM.
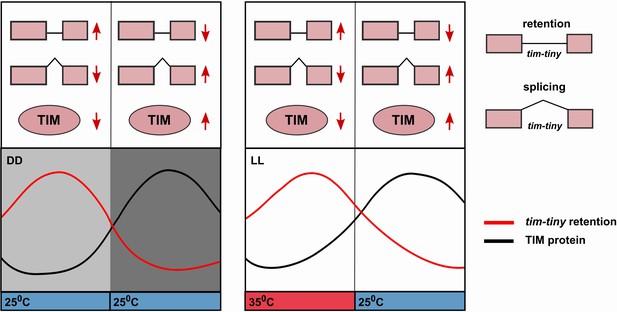
Alternative splicing of the tim-tiny intron promotes oscillations of TIM levels.
The model depicts how retention of the tim-tiny intron, which is increased upon downregulation of prp4, regulates TIM cycling. Both the circadian clock (A) and temperature cycles (B) regulate retention of the tim-tiny intron. (A) Increased retention of tim-tiny during the subjective day (light gray) in dark:dark (DD) conditions serves to decrease TIM levels and delay the accumulation of TIM in the absence of light. (B) Higher temperatures, typically associated with daytime hours, increase tim-tiny retention. Temperature cycles can maintain clock function under constant light conditions (LL), which would otherwise disrupt the clock. Entrainment by temperature appears to be driven by a reduction of TIM protein at the higher temperature (Yoshii, T., et al., 2005). We propose that under temperature cycles, retention of tim-tiny sets the levels of TIM and contributes to maintenance of the molecular clock.
Tables
Free-running locomotor behavior of flies expressing RNAi against tri-snRNP components
https://doi.org/10.7554/eLife.39821.006| Genotype | N | rhythmicity* (%) | Period (hours ± SEM) | Power (FFT ± SEM) |
|---|---|---|---|---|
| TUG; Dcr2/+ | 35 | 100% | 23.80 (0.06) | 0.11 (0.01) |
| pdfGal4, Dcr2/+ | 19 | 100% | 24.06 (0.06) | 0.10 (0.01) |
| elavGal4; Dcr2/+ | 30 | 97% | 23.59 (0.33) | 0.06 (0.03) |
| +/prp4RNAi(GD) | 16 | 100% | 23.62 (0.06) | 0.09 (0.01) |
| +/prp4RNAi(KK) | 16 | 100% | 23.32 (0.05) | 0.13 (0.01) |
| +/prp3RNAi(GD) | 10 | 100% | 23.73 (0.06) | 0.10 (0.01) |
| +/prp3RNAi(KK) | 15 | 100% | 23.45 (0.06) | 0.12 (0.01) |
| +/prp8RNAi(GD) | 15 | 100% | 23.64 (0.07) | 0.11 (0.01) |
| +/prp31RNAi(KK) | 13 | 100% | 23.35 (0.06) | 0.12 (0.04) |
| +/brr2RNAi(KK) | 16 | 100% | 23.47 (0.06) | 0.11 (0.01) |
| TUG; Dcr2 > prp4RNAi(GD) | 34 | 71%† | 26.55 (0.29)‡ | 0.09 (0.01) |
| TUG; Dcr2 > prp4RNAi(KK) | 40 | 45%† | 29.47 (0.48)‡ | 0.09 (0.01) |
| TUG; Dcr2 > prp3RNAi(GD) | 11 | 100% | 27.45 (0.49)‡ | 0.08 (0.02) |
| TUG; Dcr2 > prp3RNAi(KK) | 33 | 0%† | - | - |
| TUG; Dcr2 > prp8RNAi(GD) | 30 | 23%† | 28.00 (1.02)‡ | 0.02 (0.01)‡ |
| TUG;Dcr2 > prp31RNAi(KK) | 17 | 88% | 25.33 (0.19)‡ | 0.06 (0.01)¶ |
| TUG;Dcr2 > brr2RNAi(KK) | 24 | 0%† | - | - |
| pdfGal4, Dcr2 > prp4RNAi(GD) | 30 | 90% | 25.85 (0.32)‡ | 0.06 (0.01)|| |
| pdfGal4, Dcr2 > prp4RNAi(KK) | 38 | 97% | 24.81 (0.32)‡ | 0.09 (0.02) |
| elavGal4; Dcr2 > prp4RNAi(GD) | 27 | 52%† | 24.54 (0.14)§ | 0.03 (0.02)# |
-
* Flies with FFT value >0.01 are considered to be rhythmic.
†p < 0.001 compared to both of the heterozygous controls, by χ2 analysis.
-
‡p < 0.001 compared to both of the heterozygous controls, by Student’s t test.
§p < 0.001 compared to RNAi control but not significant (p > 0.05) compared to elavGal4; Dcr2/+ control, by
-
Student’s t test.
¶p < 0.01 compared to TUG; Dcr2/+ control but not significant (p > 0.05) compared to RNAi control, by Student’s t test.
-
||p < 0.01 compared to pdfGal4; Dcr2/+ control and p < 0.05 compared to RNAi control, by Student’s t test.
#p < 0.05 compared to RNAi control but not significant (p > 0.05) compared to elavGal4; Dcr2/+ control, by Student’s t test.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | prp4 | NA | FLYB:FBgn0027587 | |
| Gene (Drosophila melanogaster) | timeless (tim) | NA | FLYB:FBgn0014396 | |
| Gene (Drosophila melanogaster) | period (per) | NA | FLYB:FBgn0003068 | |
| Gene (Drosophila melanogaster) | prp8 | NA | FLYB:FBgn0033688 | |
| Gene (Drosophila melanogaster) | brr2 | NA | FLYB:FBgn0263599 | also known as l(3)72Ab |
| Gene (Drosophila melanogaster) | prp3 | NA | FLYB:FBgn0036915 | |
| Gene (Drosophila melanogaster) | prp31 | NA | FLYB:FBgn0036487 | |
| Strain, strain background (Drosophila melanogaster) | iso31 | from laboratory stocks | NA | |
| Genetic reagent (Drosophila melanogaster) | per01 | Bloomington Drosophila Stock Center (BDSC) | FLYB:FBal0013649 | |
| Genetic reagent (Drosophila melanogaster) | tim0 | BDSC | FLYB:FBal0035778 | |
| Genetic reagent (Drosophila melanogaster) | TUG (Tim-UAS-Gal4) | BDSC | FLYB:FBtp0011839 | |
| Genetic reagent (Drosophila melanogaster) | pdfGal4; pdfG4 | BDSC | FLYB:FBtp0011844 | |
| Genetic reagent (Drosophila melanogaster) | elavGal4; elavG4 | BDSC | BDSC:25750 | |
| Genetic reagent (Drosophila melanogaster) | GMRGal4; GMR | BDSC | FLYB:FBti0002994 | |
| Genetic reagent (Drosophila melanogaster) | prp4RNAi(GD) | Vienna Drosophila Resource Center (VDRC) | VDRC:27808 | |
| Genetic reagent (Drosophila melanogaster) | prp4RNAi(KK) | VDRC | VDRC:107042 | |
| Genetic reagent (Drosophila melanogaster) | prp8RNAi(GD) | VDRC | VDRC:18565 | |
| Genetic reagent (Drosophila melanogaster) | prp3RNAi(GD) | VDRC | VDRC:25547 | |
| Genetic reagent (Drosophila melanogaster) | prp3RNAi(KK) | VDRC | VDRC:103628 | |
| Genetic reagent (Drosophila melanogaster) | prp31RNAi(KK) | VDRC | VDRC:103721 | |
| Genetic reagent (Drosophila melanogaster) | brr2RNAi(KK) | VDRC | VDRC:110666 | |
| Genetic reagent (Drosophila melanogaster) | prp82e1 | BDSC | FLYB: FBal0190235; BDSC:25905 | |
| Genetic reagent (Drosophila melanogaster) | prp82e2 | BDSC | FLYB:FBal0190015; BDSC:25912 | |
| Genetic reagent (Drosophila melanogaster) | brr2e03171 | BDSC | FLYB:FBti0041681; BDSC:18127 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Dicer2; Dcr2 | BDSC | FLYB:FBtp0036672 | |
| Genetic reagent (Drosophila melanogaster) | UAS-tim-spliced; tim-spliced | this paper | NA | generated by the site-specific PhiC31 Integration System (Rainbow Transgenics) using the attP on the 3rd chromosome; pUAST-tim-spliced plasmid was used for injection |
| Genetic reagent (Drosophila melanogaster) | UAS-tim-retained; tim-retained | this paper | NA | generated by the site-specific PhiC31 Integration System (Rainbow Transgenics) using the attP on the 3rd chromosome; pUAST-tim- retained plasmid was used for injection |
| Genetic reagent (Drosophila melanogaster) | UAS-tim-retained+ssM; tim-retained+ ssM | this paper | NA | generated by the site-specific PhiC31 Integration System (Rainbow Transgenics) using the attP on the 3rd chromosome; pUAST-tim- retained+ssM plasmid was used for injection |
| Cell line (Drosophila melanogaster) | S2 | ATCC (Manassas, VA) | FLYB:FBtc0000181; RRID:CVCL:Z992 | |
| Antibody | guinea pig anti- PER (UP1140) | Garbe et al., 2013 | NA | 1:1000 |
| Antibody | rat anti- TIM (UPR42) | Jang et al., 2015 | NA | 1:1000 |
| Antibody | rabbit anti-PDF (HH74) | Garbe et al., 2013 | NA | 1:500 |
| Antibody | mouse anti-LaminC | Developmental Studies Hybridoma Bank (DSHB) | LC28.26 | 1:500 |
| Antibody | mouse anti-HSP70 | Sigma | Cat# H5147 | 1:5000 |
| Recombinant DNA reagent | pIZ/V5-His plasmid | ThermoFischer | Cat# V800001 | backbone |
| Recombinant DNA reagent | pBluescript-tim | lab collection | NA | tim sequence contained tim-tiny; used for subcloning |
| Recombinant DNA reagent | tim-spliced; pIZ-tim-spliced | this paper | NA | tim cDNA was subcloned into pIZ-V5 plasmids |
| Recombinant DNA reagent | tim-retained; pIZ-tim-retained | this paper | NA | tim-tiny intron was subcloned into pIZ-tim- spliced vector from pBluescript-tim plasmid |
| Recombinant DNA reagent | tim-retained+ ssM; pIZ-tim-retained+ssM | this paper | NA | generated by mutagenesis of the 5’ splice donor site of tim-tiny intron from pIZ-tim-retained plasmid |
| Recombinant DNA reagent | tim-spliced; pUAST-tim-spliced | this paper | NA | tim cDNA was subcloned from pIZ-tim- spliced into pUAST -attB vector |
| Recombinant DNA reagent | tim-retained; pUAST-tim-retained | this paper | NA | tim cDNA was subcloned from pIZ-tim-retained into pUAST-attB vector |
| Recombinant DNA reagent | tim-retained + ssM; pUAST-tim-retained+ssM | this paper | NA | tim cDNA was subcloned from pIZ-tim- retained+ssM into pUAST-attB vector |
| Sequence- based reagent | tim PP11542 (‘mRNA’) _F | ATGGACTGGTTACTAGCAACTCC | ||
| Sequence- based reagent | tim PP11542 (‘mRNA’) _R | GGTCCTCATAGGTGAGCTTGT | ||
| Sequence- based reagent | per_F | CGTCAATCC ATGGTCCCG | ||
| Sequence- based reagent | per_R | CCTGAAAGACGCGATGGTG | ||
| Sequence- based reagent | clk_F | GGATGCCAATGCCTACGAGT | ||
| Sequence- based reagent | clk_R | ACCTACGAAAGTAGCCCACG | ||
| Sequence- based reagent | prp4_F | CACAAGCAGCATCTTTGTATGG | ||
| Sequence- based reagent | prp4_R | TGTGGAGTCCCACATTCTTG | ||
| Sequence- based reagent | tim-tiny_retained_F | AAACGTGAGTTAAAGTCAACC | ||
| Sequence- based reagent | tim-tiny_retained_R | GAGAGGCACACAGCATATC | ||
| Sequence- based reagent | tim-tiny_spliced_F | CCGCTGGACAAACTCAACCTC | ||
| Sequence- based reagent | tim-tiny_spliced_R | TCGGTATCGCCGAGATCCACG | ||
| Sequence- based reagent | tim-cold_retained_F | GGCTCATGATCATTGCAGCAGC | ||
| Sequence- based reagent | tim-cold_retained_R | ATAGTGGGGCACCCGGATCTC | ||
| Sequence- based reagent | tim-cold_spliced_F | TTAAACAGCGACAATGTCTCTTTGG | ||
| Sequence- based reagent | tim-cold_spliced_R | GAATTGGATCCTCAGTGATAGTGGG | ||
| Sequence- based reagent | tim_non_spanning ('exon')_F | GAAGAACAACGATATTGTGGGAAAG | ||
| Sequence- based reagent | tim_non_spanning ('exon')_R | AGTGGGAGTTGTCAGCAAAG | ||
| Sequence- based reagent | per_retained_F | GAGGACCAGACACAGCACGG | ||
| Sequence- based reagent | per_retained_R | CGGAGGCAATTGCTCACTCGT | ||
| Sequence- based reagent | per_spliced_F | GAGGACCAGACACAGCACGG | ||
| Sequence- based reagent | per_spliced_R | TCGCGTTGATTCGAAGAATCGTT | ||
| Sequence- based reagent | rp49_F | GACGCTTCAAGGGACAGTATCTG | ||
| Sequence- based reagent | rp49_R | AAACGCGGTTCTGCATGAG | ||
| Sequence- based reagent | tim_tinySSdonorT > A_F | CTGGACAAACGAGAGTTAAAGTCAACC | ||
| Sequence- based reagent | tim_tinySSdonorT > A_R | CGGTCCCAGCTTTTTGGC | ||
| Commercial assay or kit | RNeasy Plus Mini Kit | Qiagen | Cat# 74134 | |
| Commercial assay or kit | Superscript II Reverse Transcriptase | ThermoFischer | Cat# 18064014 | |
| Commercial assay or kit | TRIzol Reagent | ThermoFischer | Cat# 15596026 | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | NEB | Cat# E0554S | |
| Commercial assay or kit | Effectene Transfection Reagent | Qiagen | Cat# 301425 | |
| Software, algorithm | Graphpad Prism v7 | Graphpad Software | https://www.graphpad.com/ | |
| Software, algorithm | JTK_CYCLE v3 | Hughes et al., 2010 | NA | |
| Software, algorithm | ImageJ | NIH | https://imagej.nih.gov/ij/ | |
| Software, algorithm | ClockLab Software | Actimetrics (Wilmette, IL) | https://actimetrics.com/products/clocklab |
Additional files
-
Supplementary file 1
DAVID pathway enrichment analysis of all differentially expressed genes upon prp4 knockdown
- https://doi.org/10.7554/eLife.39821.014
-
Supplementary file 2
List of differentially expressed genes upon prp4 knockdown
- https://doi.org/10.7554/eLife.39821.015
-
Supplementary file 3
Differentially spliced events upon prp4 knockdown identified with CASH
- https://doi.org/10.7554/eLife.39821.016
-
Supplementary file 4
Differentially spliced isoforms upon prp4 knockdown identified with Cufflinks-2.2 pipeline
- https://doi.org/10.7554/eLife.39821.017
-
Supplementary file 5
Calculation of the ratio of tim-tiny retaining isoforms to all other isoforms using Cufflinks-2.2 output
- https://doi.org/10.7554/eLife.39821.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39821.019





