BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands
Figures
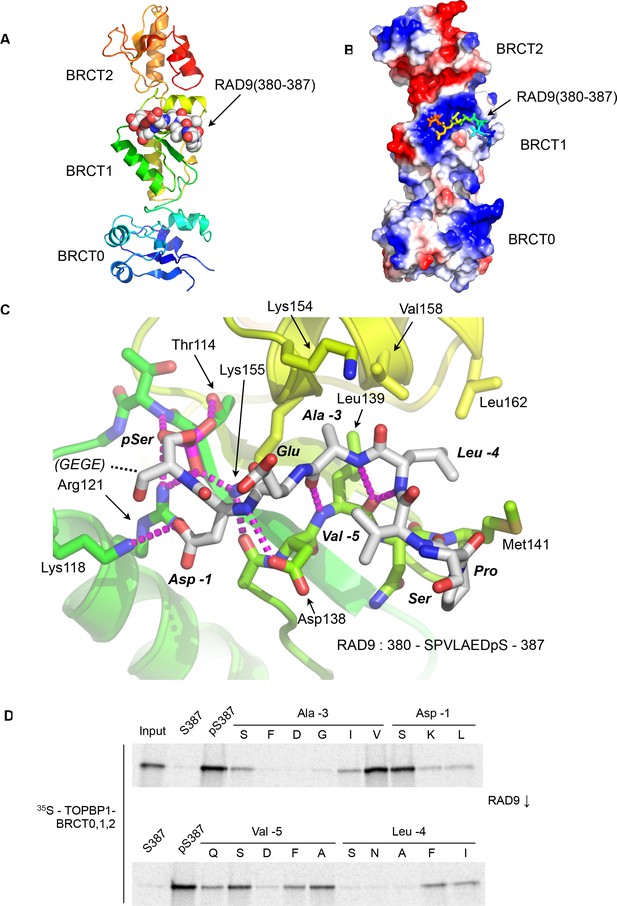
Crystal structure of the TOPBP1 – RAD9 interaction.
(A) Secondary structure cartoon (rainbow coloured N:blue → C:red) of the BRCT0,1,2 module of chicken TOPBP1 bound to the C-terminal tail of human RAD9 (CPK model) phosphorylated on Ser387. (B) The RAD9-pS387 peptide binds to a positively charged patch (blue) on BRCT1 of TOPBP1 (C) Close-up of the TOPBP1 - RAD9 interaction. The negatively charged phosphate group of RAD9-pSer387 makes multiple hydrogen binding interactions with residues that are strongly topologically conserved in all phosphopeptide-binding BRCT domains. An additional polar interaction is provided by RAD9-Asp386. The consecutive hydrophobic side chains of RAD9-Leu383 and Ala384 insert into a pocket in TOPBP1-BRCT1, enabled by their main chains packing against the side chain of Val382, in a tight turn conformation. (D) Sub-site preferences of RAD9 tail binding to TOPBP1-BRCT1. Biotinylated peptides based on the C-terminal residues of HsRAD9 with or without phosphorylation on Ser387, and pS387 peptides with single point mutations in positions −1, –3, −4 and −5 relative to the phosphorylated residue, were used to pull-down radiolabelled in vitro translated TOPBP1-BRCT0,1,2 (see Materials and methods), with the relative yields of bound protein in the autoradiographs reflecting which amino acids can be accommodated at the different positions in the bound peptide sequence (see text).
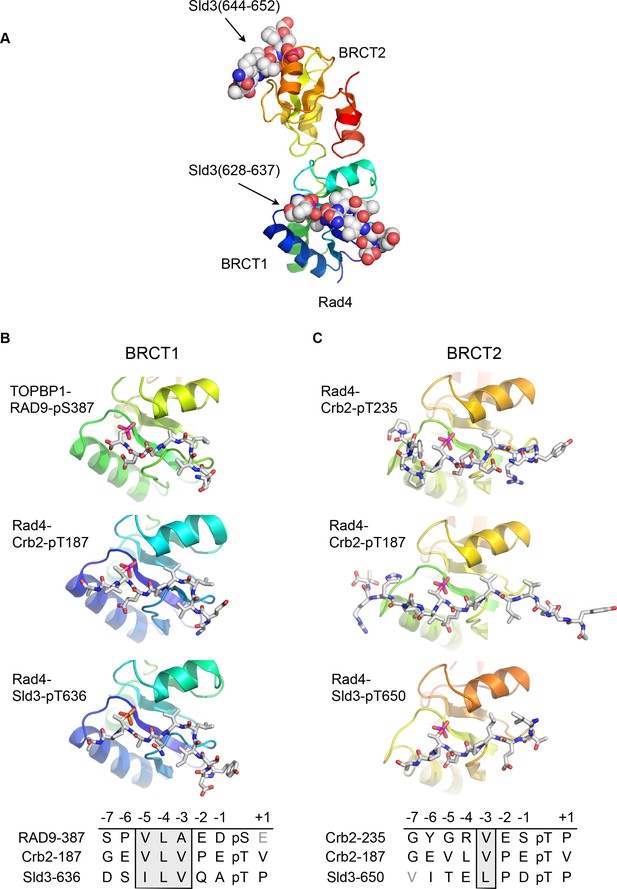
Conserved conformations of BRCT1 and BRCT2 ligands.
(A) Crystal structure of the BRCT1,2 module of Rad4, the fission yeast homologue of TOPBP1, bound simultaneously to peptides from Sld3, the fission yeast homologue of Treslin, phosphorylated on Thr636 and on Thr650. (B) Montage of crystal structures of phosphopeptide complexes with BRCT1 of TOPBP1 or Rad4 – the sequences of the peptides is shown below. The other BRCT domains present in the crystals are omitted for clarity. All three examples share a common tight turn conformation for the −5, –4 and −3 region, as described above for TOPBP1-RAD9. (C) As B, but for BRCT2. Unlike BRCT1, the −5, –4, −3 regions of the bound peptides have an extended backbone conformation, in which only the hydrophobic side chain at position −3 binds into a pocket on the BRCT domain.
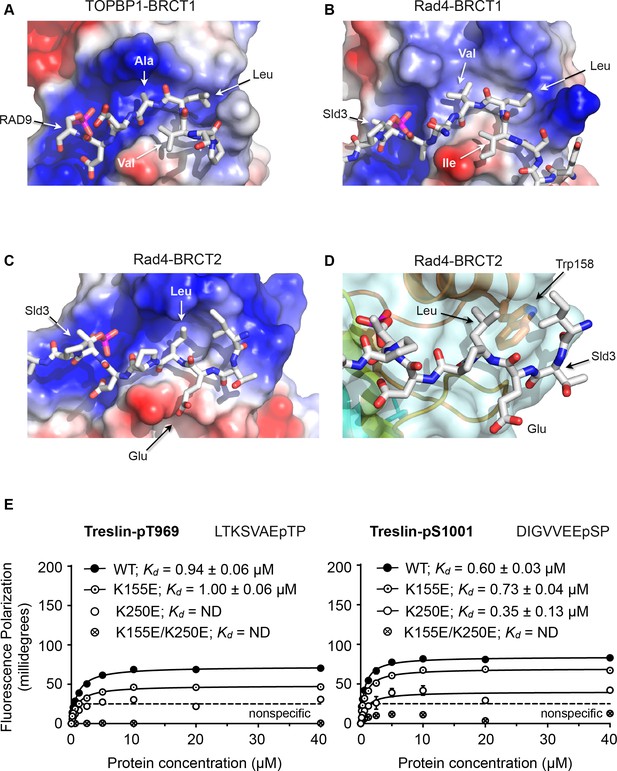
Structural basis of BRCT domain conformational preferences
(A) Detail of the RAD9-pSer387 peptide bound to TOPBP1-BRCT1, showing the tight turn conformation adopted by the consecutive hydrophobic residues at positions −5, –4 and −3 relative to the phosphorylated residue, that positions the side chains of the −4 and −3 residues in a pocket on the BRCT domain. (B) As A. but for the Sld3-pT650 peptide bound to Rad4-BRCT1. (C) Detail of the Sld3-pT650 peptide bound to Rad4-BRCT2. Unlike the BRCT1 ligand phosphopeptides, residues −3 and −4 have an extended main chain conformation, so that only the residue at −3 interacts with the hydrophobic pocket on the BRCT domain, so that the −4 position can accommodate a large polar amino acid such as glutamic acid. (D) The pocket in BRCT2 is constricted relative to that in BRCT1, by the presence of a tryptophan residue, which is topologically conserved in BRCT2 domains but absent from BRCT1. (E) Fluorescence polarisation assay of Treslin-derived phosphopeptides binding to the TOPBP1-BRCT0,1,2 segment. The Treslin-pT969 peptide, which has a large hydrophilic residue at −5 binds preferentially to BRCT2, whereas the Treslin-p1001 peptide with a glycine at −5 binds with comparable affinity to either. Both Treslin phosphopeptides have hydrophobic residues at the −3 positions.
-
Figure 3—source data 1
Fluorescence polarisation titration of Treslin binding to TOPBP1.
- https://doi.org/10.7554/eLife.39979.005
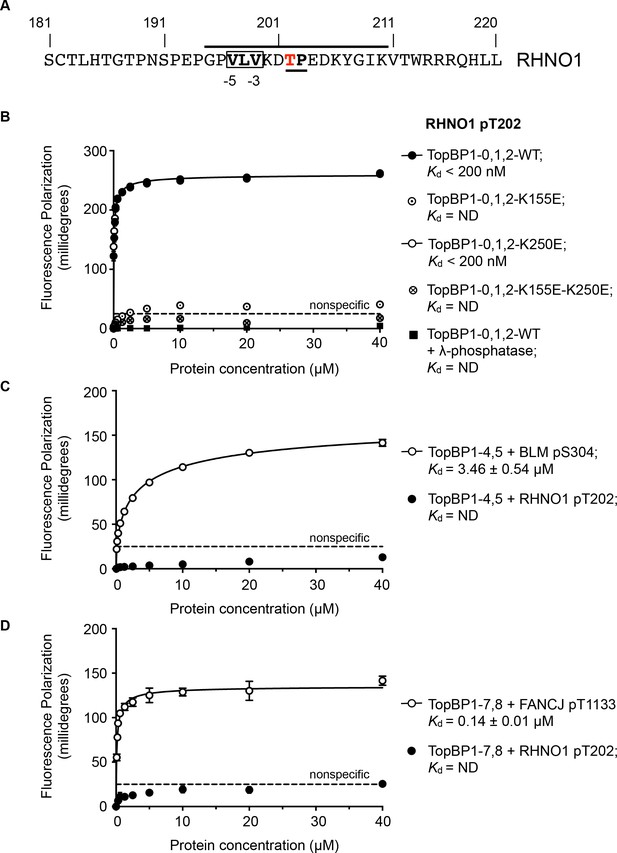
TOPBP1-binding site in RHNO1.
(A) The C-terminal half of the 9-1-1 and TOPBP1-interacting scaffold protein RHNO1, contains a sequence motif with a potential phosphorylation site at Thr202, that corresponds closely to the consensus for preferential binding to Rad4/TOPBP1-BRCT1 or BRCT2 (B) Fluorescence polarisation assay of a RHNO1-derived phosphopeptide binding to the TOPBP1-BRCT0,1,2 segment. The RHNO1-pT202 peptide binds with high affinity to the wild-type BRCT0,1,2 construct, but fails to bind when dephosphorylated by λ-phosphatase. High affinity binding is lost in the presence of disruptive mutations in the phosphate-binding site of BRCT1, but is unaffected by comparable mutations in BRCT2. (C) In contrast to a documented phosphopeptide derived from BLM (Blackford et al., 2015), the RHNO1-pT202 peptide shows no affinity for the TOPBP1-BRCT4,5 segment. (D) As C – RHNO1-pT202 shows no affinity for the TOPBP1-BRCT7,8, unlike a documented phosphopeptide derived from FANCJ (Gong et al., 2010).
-
Figure 4—source data 1
Fluorescence polarisation titration of RHNO1 binding to TOPBP1.
- https://doi.org/10.7554/eLife.39979.008
-
Figure 4—source data 2
Fluorescence polarisation titration of RHNO1 and BLM binding to TOPBP1.
- https://doi.org/10.7554/eLife.39979.009
-
Figure 4—source data 3
Fluorescence polarisation titration of RHNO1 and FANCJ binding to TOPBP1.
- https://doi.org/10.7554/eLife.39979.010

Conservation of the TOPBP1-interacting phosphorylation site in RHINO homologues.
Alignment of C-terminal RHINO sequences showing very strong conservation of the TOPBP1-interacting motif identified in human RHNO1 across a broad range of animals. Sequences are from humans (Homo sapiens), rodents (Rattus norvegicus), ruminants (Ovis aries), fish (Salmo salar), birds (Gallus gallus), cephalopods (Octopus bimaculoides), amphibians (Xenopus laevis), reptiles (Chelonia mydas) and bivalves (Mizuhopecten yessoensis).
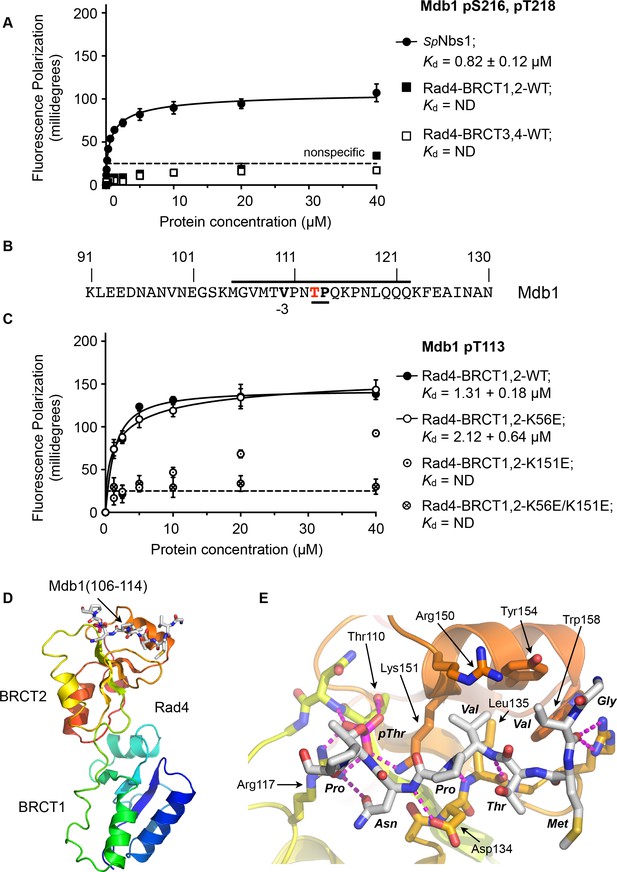
Rad4-binding site in Mdb1.
(A) Fluorescence polarisation assay showing that a fluorescent peptide (Flu-GYGVEGDHS(pS)D(pT)EEEVVS) incorporating the single SDT site in Mdb1, the fission yeast homologue of MDC1, does not interact with Rad4-BRCT3,4 as has been suggested for the SDT sites in human MDC1 with the homologous TOPBP1-BRCT4,5 (Leung et al., 2013; Wang et al., 2011). The Mdb1-SDT site peptide does, however, interact with high-affinity with the S. pombe homologue of NBS1. (B) Mdb1 contains a sequence motif with a potential phosphorylation site at Thr 113 that corresponds closely to the consensus for binding to Rad4/TOPBP1-BRCT2. (C) Fluorescence polarisation assay of an Mdb1-derived phosphopeptide binding to the Rad4-BRCT1,2 segment. The Mdb1-pT113 peptide binds with high affinity to the wild-type BRCT1,2 but only weakly when a disruptive mutation is introduced into the phosphate-binding site of BRCT2, but is largely unaffected by comparable mutations in BRCT1. All detectable bindings are lost when both BRCT domains are mutated. (D) Crystal structure of Mdb1-pT113 peptide bound to the BRCT2 domain within the Rad4-BRCT1,2 module. (E) Closeup of interactions. As in other complexes with BRCT2, the single hydrophobic residue at −3 relative to the phosphorylated residue binds into the hydrophobic pocket, with the main chain for the −2, –3 and −4 residues adopting an extended conformation, rather than the tight turn seen in interactions with BRCT1.
-
Figure 5—source data 1
Fluorescence polarisation titration of Mdb1 STD sites binding to Rad4 or SpNbs1.
- https://doi.org/10.7554/eLife.39979.014
-
Figure 5—source data 2
Fluorescence polarisation titration of Mdb1 binding to Rad4.
- https://doi.org/10.7554/eLife.39979.015
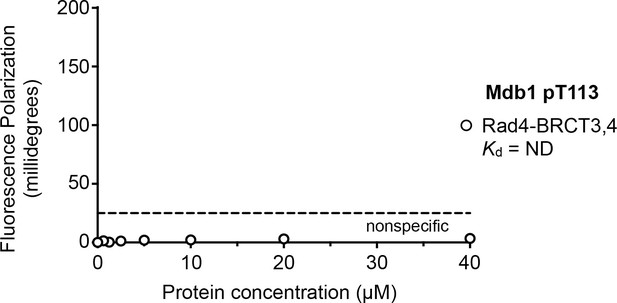
Fluorescence polarisation assay of an Mdb1-derived phosphopeptide binding to the Rad4-BRCT3,4 segment.
In contrast to its high-affinity interaction with the Rad4-BRCT1,2 segment, no interaction is observed between the Mdb1 pT113 motif and Rad4-BRCT3,4.
-
Figure 5—figure supplement 1—source data 1
Fluorescence polarisation titration of Mdb1 binding to Rad4.
- https://doi.org/10.7554/eLife.39979.013
Additional files
-
Supplementary file 1
Crystallographic data collection and refinement statistics.
- https://doi.org/10.7554/eLife.39979.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39979.018





