Comparisons between the ON- and OFF-edge motion pathways in the Drosophila brain
Figures
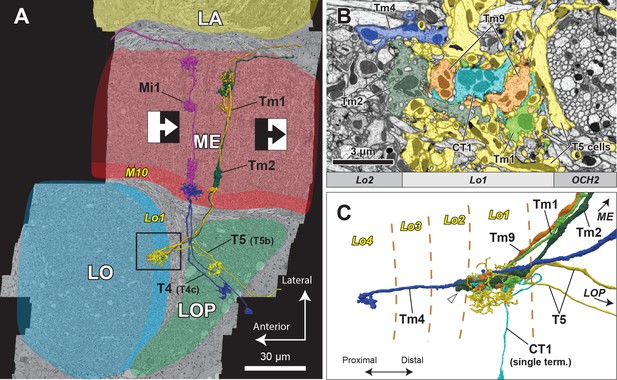
Horizontal section of the four optic lobe neuropils and their reconstructed neurons.
(A) In the ON-edge pathway, the T4 cells receive inputs at the tenth layer of the medulla (M10) from upstream cells, whereas in the OFF-edge pathway, the T5 cells receive inputs at the first layer of the lobula (Lo1), before projecting to the lobula plate. Here, in a horizontal re-sliced section of the dataset, Mi1 and Tm1/Tm2 cells are shown as representative input neurons to the T4 and T5 cells. LA, lamina; ME, medulla; LO, lobula; LOP, lobula plate. (B) Axon terminals of Tm cells and T5 cells in Lo1. The area roughly corresponds to the boxed inset in (A). T5 cell dendrites (in yellow) demarcate the Lo1 layer. Terminals of Tm1, Tm2, Tm9, and CT1 neurons form packed columnar units and have extensive synaptic contacts with T5 cells. Tm4 cells form no part of these columnar units but run between the columns, and also have synapses with T5 cells in Lo1. (C) 3D reconstruction of the neurons. One representative T5 cell (yellow) is shown along with Tm cells and a CT1 terminal. The proximal tip of the axon terminal of Tm2 (arrowhead) extends slightly beyond the boundary between Lo2 and Lo1. Tm4 extends its axon further and terminates in Lo4 (Fischbach and Dittrich, 1989).
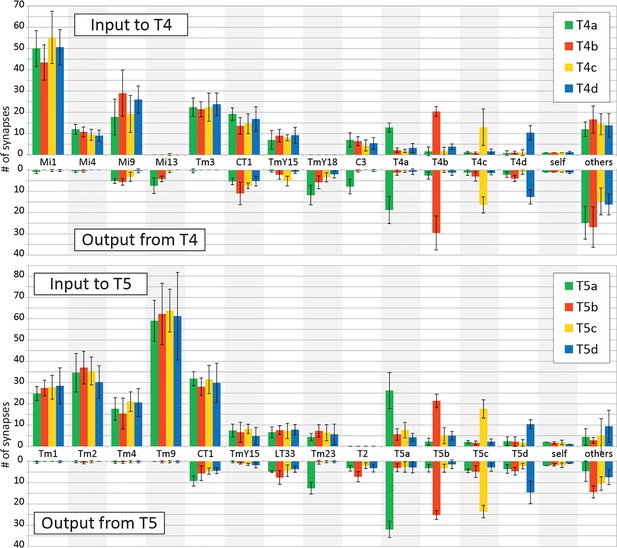
Input and output synapses of T4 and T5 cells in the medulla and lobula.
Histograms indicating the average numbers of synapses per connection for a single T4 or T5 cell, mean (+SD) for five representative cells selected and completely traced for each T4/T5 subtype. Only connections with ≥ 3 contacts are independently shown in the graphs; weaker connections are combined in ‘others’. All synaptic sites were manually verified, and their pre- and postsynaptic partners were then also exhaustively identified.
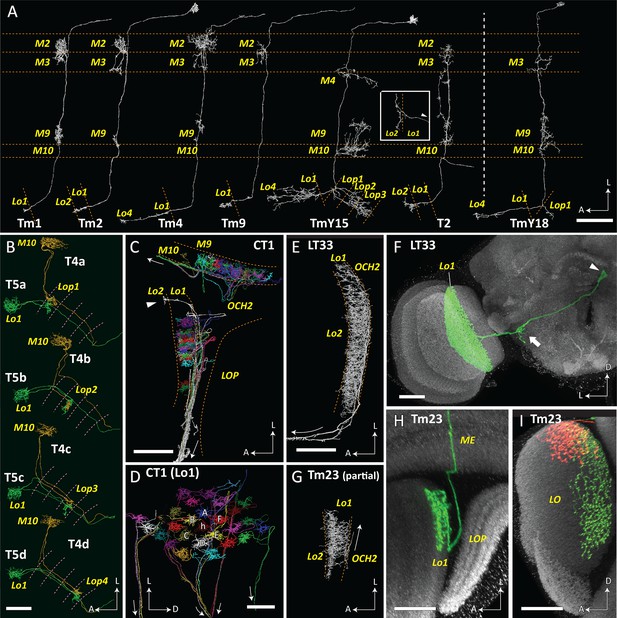
T4, T5 and their synaptic partner neurons.
(A) T5 forms synaptic contacts with Tm1, 2, 4, 9, TmY15 and T2 cells. T4 is presynaptic to a newly identified TmY18 cell in M10. These synaptic partners of T5 and T4 have cell bodies in the medulla cortex distal to stratum M1, except T2 has a soma between the medulla and lobula. Tm and T2 cells receive inputs mainly in the medulla and project to the lobula, whereas TmY cells project to both the lobula and the lobula plate. All reconstructed cell profiles closely capture the profiles of those reported from Golgi impregnation (Fischbach and Dittrich, 1989) except the lobula terminal of T2 (inset), shown here from a different angle (lateral view, arrowhead indicates the outgoing fiber connected to the medulla and cell body). (B) Paired T4 and T5 cells. T4/5a, b, c, and d cell pairs project to the lobula plate, to strata Lop1, 2, 3 and 4, respectively. Their cell bodies are located in the lobula plate cortex, posterior to the lobula plate. (C, D) Parts of CT1 reconstructed in both the medulla and lobula reveal the morphology of its color-coded columnar terminals. Fibers connecting the terminals enter the second optic chiasm (OCH2) before exiting the optic neuropils (trajectories shown by arrows). Most branches terminate in medulla stratum M10 or lobula stratum Lo1, but a few terminals project deeper, into Lo2 (arrowhead in (C)). Columnar terminals in Lo1 corresponding to a home (h) column and the surrounding six columns (A–F) are indicated in (D). (E) A partial reconstruction of LT33, a novel neuron in lobula stratum Lo1. (F) Optic lobe and central brain projection pattern of an LT33 cell visualized using multicolor stochastic labeling (MCFO; see Materials and methods). The arrowhead indicates the cell body, and the arrow shows the site of branching in the posterior lateral protocerebrum. (G) Partial reconstruction of Tm23 in stratum Lo1. (H, I) Tm23 cells and their projection patterns in the optic lobe visualized using MCFO. Images show two different views generated from the same confocal stack. Images in (F), (H), and (I) show resampled views generated from confocal stacks using Vaa3D. For clarity, the images in (F) and (H) were segmented to omit additional labeled cells (distinguished by labeling color and position) that are also present in the displayed volume. In (I), red and green signals each represent single Tm23 cells. Scale bars: (A) 20 μm; (B), (C), (E), (G) 10 μm; (D) 5 μm; (F), (H), (I) 20 μm.
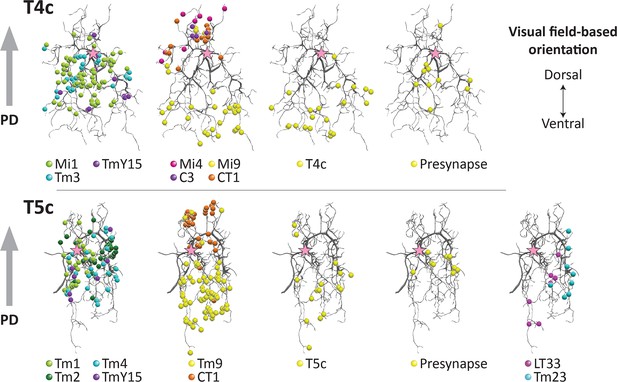
Distribution of synaptic sites over T4 and T5 dendritic arbors.
Proofread synapses are plotted on the dendritic arbors of a T4c (top panels) and T5c (bottom panels) cell. Puncta are postsynaptic sites (inputs to the T4/T5 cells) except those shown as ‘Presynapse’, which illustrate the positions of T-bar, output sites. Both T4c and T5c detect upward motion, and other subtypes of T4 and T5 cells demonstrate similar distribution patterns(not shown). Pink stars indicate the first branch point of the arbor. PD, preferred direction.
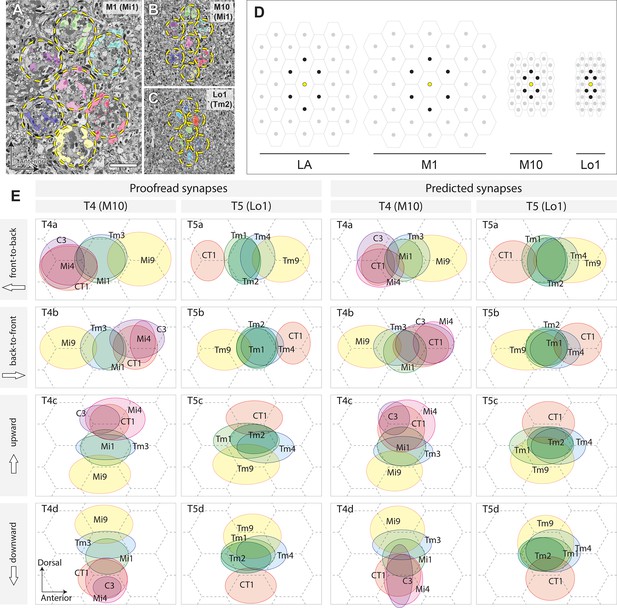
Columnar units in the optic neuropils and spatial distribution of synaptic inputs to T4 and T5 cell arbors.
(A) Retinotopic columnar terminals of Mi1 cells in stratum M1 (one center column and six surrounding columns). (B) The same subset of neurons in the proximal medulla (M10). Retinotopy of the columns is preserved in the distal lobula too. (C) The distribution of Tm2 axon terminals in layer Lo1. Locations of the columns are indicated by the broken circles. (D) Relative sizes and orientations of columnar units in different levels of the optic lobe. On the surface of the compound eye, ommatidial lenses are arranged in a regular hexagonal array. The arrangement of the lamina (LA) cartridges inherits the arrangement of the ommatidia, but is stretched in the D–V axis. The size of the column becomes larger in M1, where the lamina neurons make synapses with a number of medulla columnar neurons, including Mi and Tm cells. In M10, the size of the column becomes much smaller, and each column is elongated in the D–V axis. In the lobula, the column becomes yet smaller, and shortened in the A–P axis, making the column array triangular, rather than hexagonal. Center columns are in yellow, and six surrounding columns in black. (E) To normalize synaptic distributions on T4 and T5 dendrites, columns in M10 and Lo1 are assumed to be arranged in a regular hexagonal array, reflecting the arrangement of ommatidia. Input neurons to representative T4 and T5 cells were identified by the cell types and columns that they occupy, and their inputs were mapped onto the hexagonal grid by weighing synapses. The centers of the ellipses represent the averaged input sites, with the width and height corresponding to the standard deviations of inputs along the x- and y-axes. The left two columns are plotted using proofread synapses, whereas in the right columns, predicted (non-proofread) synapses were used. The average of all inputs was designated as the center point, which corresponds to the center of a column. The A–P and D–V axes correspond to the directions in the visual field, not to the body axis. The dimensions of the receptive fields are much more accurate, and are based on many more reconstructed neurons, than those reported from the manual annotation of ssTEM series in Shinomiya et al. (2014). The synapses are also more comprehensively annotated, and the synapse annotation is much more precise. See the main text for further detail. Scale bar: (A–C) 5 μm.
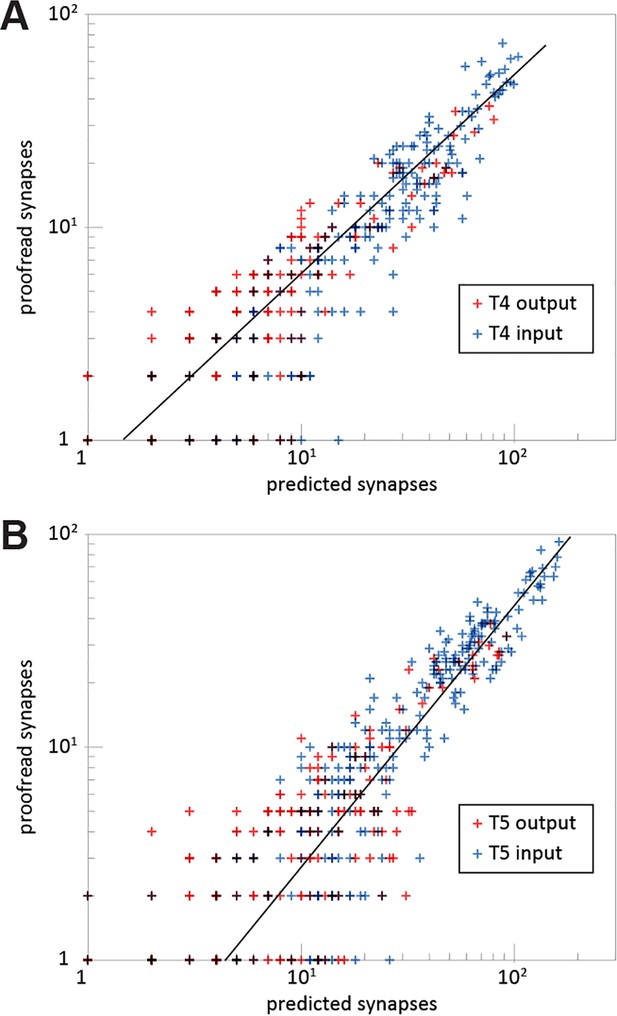
Comparison between the numbers of predicted and proofread synapses per connection.
Numbers of synapses between T4/T5 cells and their direct synaptic partners. Synapses were counted for individual T4 and T5 cells, whereas synaptic partners are grouped by cell type (Mi1, Tm1, CT1…). Both axes are plotted on a logarithmic scale, and the x-axis corresponds to the number of predicted synapses (those not proofread), whereas the y-axis corresponds to the number of proofread synapses. 260 inputs to T4 and 260 outputs from T4 are plotted in (A), and 280 inputs to T5 and 280 outputs from T5 are plotted on (B); pairs of neuron types lacking synaptic partnerships are omitted. The regression line in (A) is y = 0.5304x – 0.1835 (R²=0.880), and in (B) is y = 0.4585x – 0.9519 (R²=0.909).
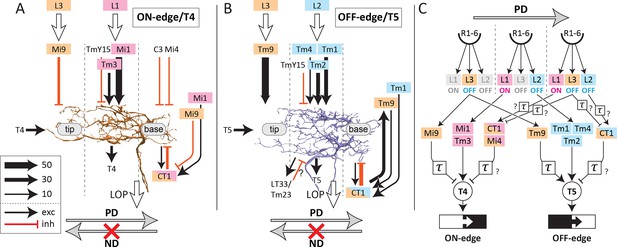
Schematic diagram of inputs onto and outputs from T4 and T5 dendrites.
Major input and output neurons of the dendritic arbors of T4 (A) and T5 (B), ranked according to synapse number (key), with dotted lines indicating the approximate boundaries of the corresponding medulla/lobula columns. (C) Schematic circuit model of the ON-edge and OFF-edge pathways. A temporal delay is indicated by τ. See the text for details. Medulla neurons receiving inputs from L1, L2, and L3 cells are color-coded accordingly. PD, preferred direction; ND, non-preferred direction.
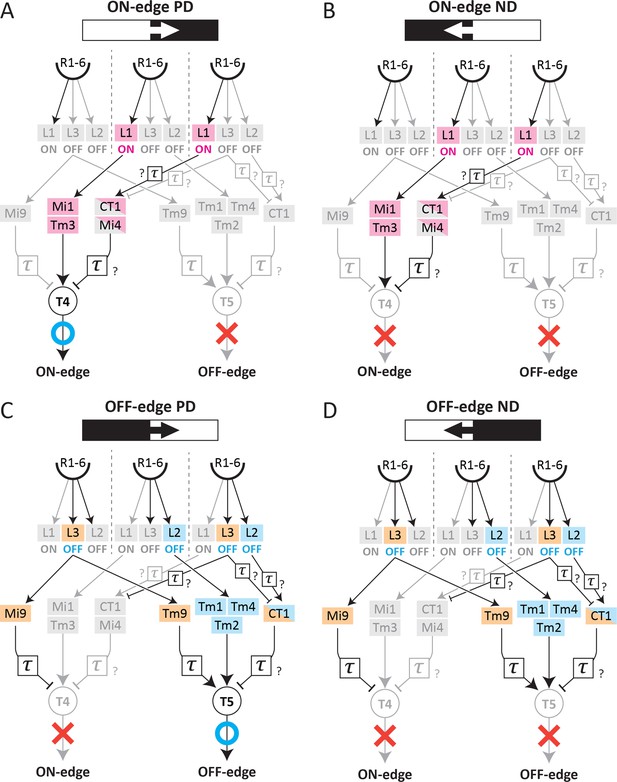
Circuit diagrams for ON- and OFF-edge models that signal motion.
The motion circuit neurons activated by signals from ON-edge motion in either the preferred (A), or non-preferred (B) directions, and by OFF-edge motion in either the preferred (C), or non-preferred (D) directions. T4 and T5 cell signal motion only in the preferred directions (A, C).
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40025.010





