Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation
Figures
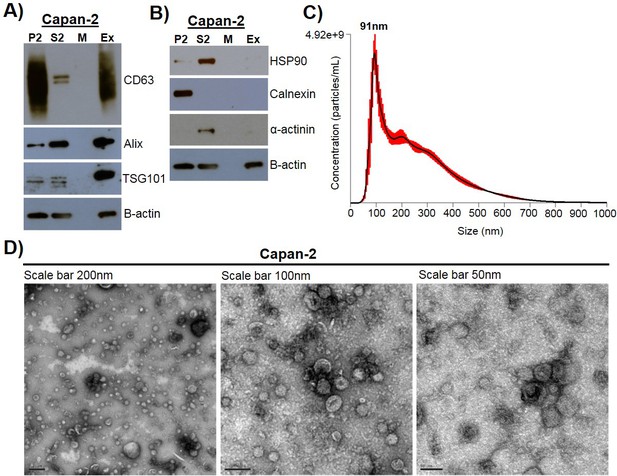
Exosome isolation, validation, and characterization.
(A) Western blot analysis of common exosomal marker proteins CD63, Alix, and TSG10 found in exosomes isolated from Capan-2 cells. (B) Western blot analysis of proteins HSP90, Calnexin, and α-actinin, expected to be underrepresented in exosomes. Equivalent amounts of proteins from P2 (ER and mitochondria), S2 (cytoplasm), M (media), and Ex (exosome) fractions derived from the Capan-2 exosome isolation process were loaded into gel for the analysis. (C) Nanoparticle Tracking Analysis of ‘crude’ Capan-2 cell exosomes. Data represent average size per concentration (black line) ± standard error of the mean (red bars) of three measurements from one exosome preparation. Exosome size is centered on 91 nm with a mean size of 250.3 nm. Finite Track Length Analysis (FTLA) was used for size determination. (D) Representative TEM images of exosomes isolated from Capan-2 cells shown at three different scales confirm expected cup-shaped morphology of vesicles.
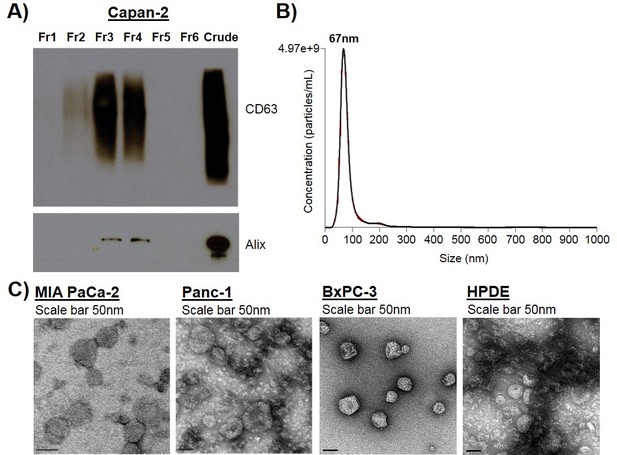
Exosome isolation, validation, and characterization of additional cell types.
(A) Western blot analysis of common exosomal marker proteins CD63 and Alix found in exosomes isolated from Capan-2 cells. Crude exosomes were isolated from the ultrafiltration-ultracentrifugation method. Crude exosomes were further purified using a sucrose density gradient to produce the six fractions (Fr1 to Fr6). Exosome marker proteins were identified primarily in fractions 3 and 4 in addition to the Crude exosome fraction. Fr3 was collected and used in the cell transformation assay as ‘pure’ exosome sample. (B) Nanoparticle Tracking Analysis of ‘Fr3’ Capan-2 cell exosomes. Data represent average size per concentration (black line) ± standard error of the mean (red bars) of three measurements from one exosome preparation. Exosome size is centered on 67 nm with a mean size of 83.5 nm. Finite Track Length Analysis (FTLA) was used for size determination. (C) TEM images of exosomes isolated from MIA PaCa-2, Panc-1, BxPC-3, and HPDE cells, scale bar is 50 nm in size. Images confirm expected cup-shaped morphology of exosomes.
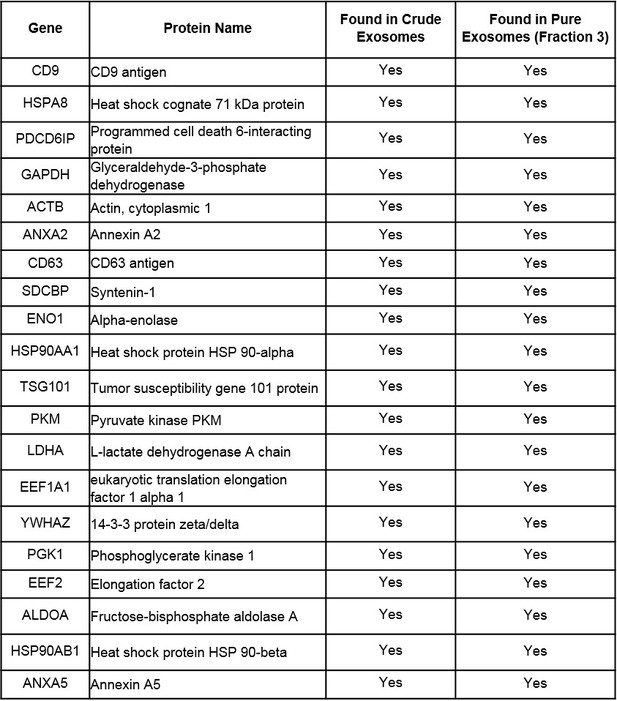
Twenty most common proteins found in exosomes according to ExoCarta database identified by mass spectrometry analysis in ‘crude’ Capan-2 exosome sample and ‘pure’ Capan-2 exosome sample (Fraction three from sucrose density gradient).
https://doi.org/10.7554/eLife.40226.004-
Figure 1—figure supplement 2—source data 1
Relates to Figure 1—figure supplement 2.
Proteins found by mass spectrometry in crude exosomes isolated from Capan-2 cells by a combined –ultrafiltration-ultracentrifugation method. PSM = peptide spectral matches.
- https://doi.org/10.7554/eLife.40226.005
-
Figure 1—figure supplement 2—source data 2
Relates to Figure 1—figure supplement 2.
Proteins found by mass spectrometry in exosomes isolated from Capan-2 cells by a combined ultrafiltration-ultracentrifugation method and further purified using sucrose density gradient separation. Only Fraction 3 was collected and analyzed further. PSM = peptide spectral matches.
- https://doi.org/10.7554/eLife.40226.006
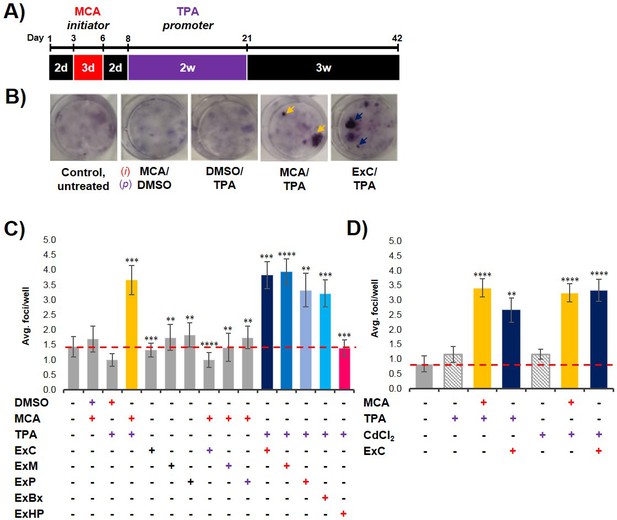
Pancreatic cancer cell exosomes function as an initiator in malignant cell transformation.
(A) Two-stage cell transformation assay shown. NIH/3T3 cells were treated with a tumor initiator for 3 days (Days 3–6) and the tumor promoter for 2 weeks (Days 8–21). After 42 days, cells are fixed with methanol and stained with Crystal Violet for malignant foci counting. (B) Representative images of stained cells showing foci formation (arrows) from untreated cells and cells treated with MCA/DMSO, DMSO/TPA, MCA/TPA, or Capan-2 exosomes (ExC)/TPA (initiator/promoter). (C) Quantification of foci formed at the end of cell transformation assays. The average foci/well were determined via double-blind counting as described in Materials and methods. The red dashed line represents the established level of background foci present in untreated cells. Initiator/promoter treatments resulting in increased foci formation above background include MCA/TPA (p=0.008) and all cancer cell-derived exosomes: ExC/TPA (p=0.0002), ExM/TPA (p<0.0001), ExP/TPA (p=0.007), and ExBx/TPA (p=0.0003). Bars shown in gray represent controls that did not result in foci formation above background. Bar shown in pink shows results from normal cell (HPDE) exosome/TPA treatment (p=0.0004). (D) Quantification of foci formed after use of a different promoter, CdCl2. CdCl2 acts as a promoter leading to increased foci formation above background when used with the initiators MCA (p<0.0001) or Capan-2 exosomes (ExC) (p<0.0001). Asterisks indicate significant differences from either control treatment or MCA/TPA treatment as determined by unpaired, two-tailed t-test with Welch’s correction (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). Red (+)=initiator used; purple (+)=promoter used.
-
Figure 2—source data 1
Relates to Figure 2.
Quantification of foci formed from two-stage cell transformation assays shown in Figure 2C.
- https://doi.org/10.7554/eLife.40226.012
-
Figure 2—source data 2
Relates to Figure 2.
Quantification of foci formed from two-stage cell transformation assays shown in Figure 2D.
- https://doi.org/10.7554/eLife.40226.013
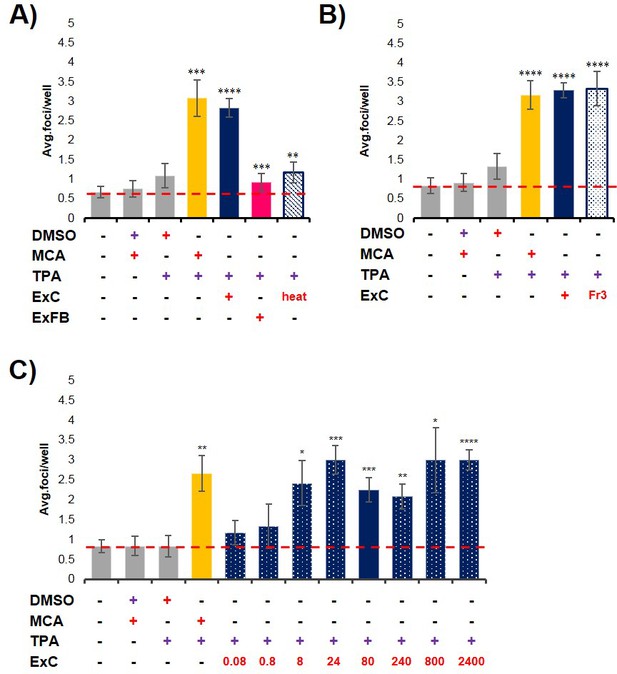
Characterization of initiation activity of pancreatic cancer cell exosomes.
(A) Quantification of foci formed at the end of cell transformation assays, foci were counted as described in Materials and methods. The red dashed line represents the established level of background foci present in untreated cells. Initiator/promoter treatments resulting in increased foci formation above background include MCA/TPA (p=0.008) and ExC/TPA (p=0.0002), as previously shown. Treatment of cells with primary fibroblasts exosomes (ExFB) (initiator)/TPA (promoter) did not result in increased cell transformation (pink bar, p=0.0007). Treatment of cells with ExC heated exosomes (initiator)/TPA (promoter) did not result in increased cell transformation (dashed blue bar, p=0.0024). (B) Treatment of cells with ‘pure’ ExC exosomes (fraction three from density sucrose gradient fractionation) as an initiator and TPA as a promoter did result in increased cell transformation above background (dotted blue bar, p<0.0001). (C) Dose-response studies. ExC exosomes were tested in the assay at a measured protein concentration ranging from 0.08 ng/mL to 2400 ng/mL (concentrations in ng/mL are shown in red). Asterisks indicate significant differences from either control treatment or MCA/TPA treatment as determined by unpaired, two-tailed t-test with Welch’s correction (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). Red (+)=initiator used; purple (+)=promoter used.
-
Figure 2—figure supplement 1—source data 1
Relates to Figure 2—figure supplement 1.
Quantification of foci formed from two-stage cell transformation assays shown in Figure 2—figure supplement 1A.
- https://doi.org/10.7554/eLife.40226.009
-
Figure 2—figure supplement 1—source data 2
Relates to Figure 2—figure supplement 1.
Quantification of foci formed from two-stage cell transformation assays shown in Figure 2—figure supplement 1B.
- https://doi.org/10.7554/eLife.40226.010
-
Figure 2—figure supplement 1—source data 3
Relates to Figure 2—figure supplement 1.
Quantification of foci formed from two-stage cell transformation assays shown in Figure 2—figure supplement 1C.
- https://doi.org/10.7554/eLife.40226.011
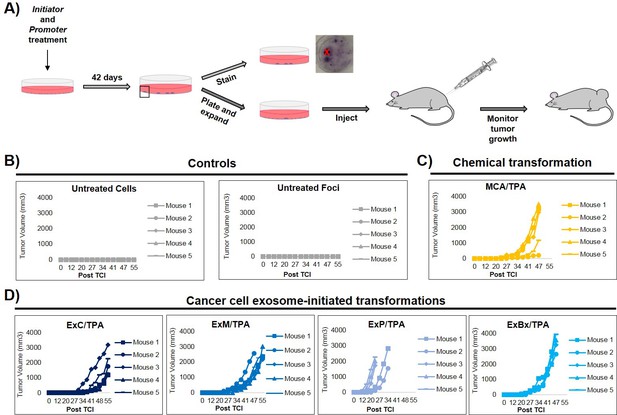
In vivo studies confirm the fully transformed form of cancer cell exosome-initiated cells.
(A) In vivo assay. NIH/3T3 cells are treated with an initiator and promoter according to the cell transformation assay (42 days total). At the end of the transformation experiment, prior to methanol fixation and staining with crystal violet, foci were isolated, expanded, and established as a transformed cell line. Transformed cells are then subcutaneously injected into mice to monitor for tumor formation. Tumor growth was tracked by measuring tumor volume 2x/week for up to 55 days post injection (Post TCI) or until tumor size exceeded maximum limit. (B) Control mice include injection of untreated NIH/3T3 cells or background foci formed in untreated NIH/3T3 cells. Cells were injected at a concentration of 1 × 106 cells. Injection of untreated cells never resulted in tumor growth; injection of background foci from untreated cells resulted in tumor growth in 6 out of 15 total mice (see figure supplements for additional mice). (C) Results from injections of chemically transformed cells (MCA = initiator/TPA = promoter). Transformed cells were injected at a concentration of 1 × 106 cells. Tumor growth was observed in all mice (n = 5) from three independent experiments (see figure supplements for additional mice). (D) Results from injections of cancer cell exosome-initiated transformed cells; exosomes from four cancer cell lines, Capan-2 (ExC), MIA PaCa-2 (ExM), Panc-1 (ExP), and BxPC-3 (ExBx), were used as an initiator with the promoter TPA. Transformed cells were injected at a concentration of 1 × 106 cells. Tumor growth was observed in all mice (n = 5) for each treatment (see figure supplements for additional mice).
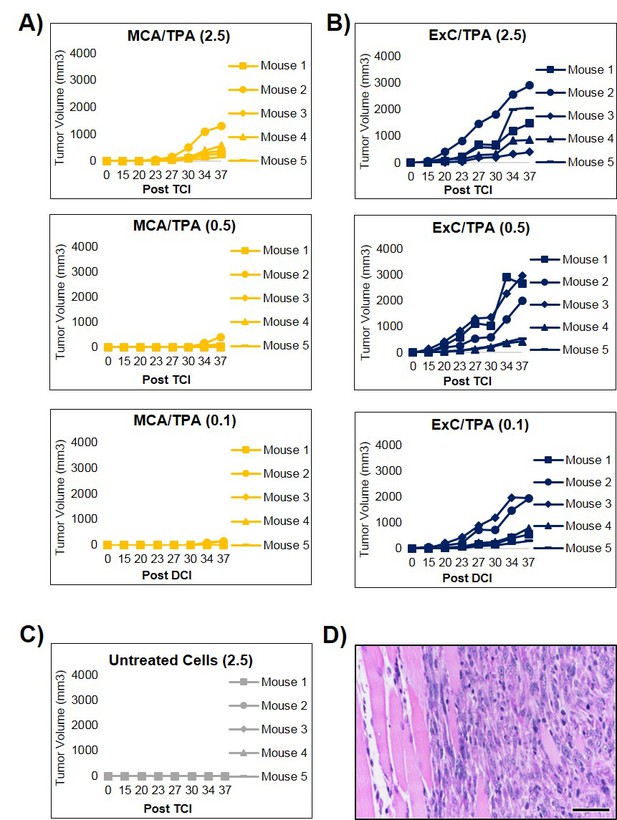
Additional in vivo studies of transformed cells.
(A–B) MCA/TPA and Capan-2 exosome (ExC)/TPA transformed cells were each subcutaneously injected into NSG (NOD scid gamma) mice at three concentrations, 2.5 × 106 (2.5), 0.5 × 106 (0.5), and 0.1 × 106 (0.1) cells, to determine sufficient cell density for tumor formation (n = 5 mice for each concentration). Tumor growth was tracked by measuring tumor volume 2x/week for 37 days post injection (Post TCI). (C) Additional replicates of control mice injected with untreated NIH/3T3 cells at a concentration of 2.5 × 106 cells. (D) Histological analysis of one representative tumor formed in mice, confirming that the tumors are fibrosarcomas, scale bar = 50 μm in size.
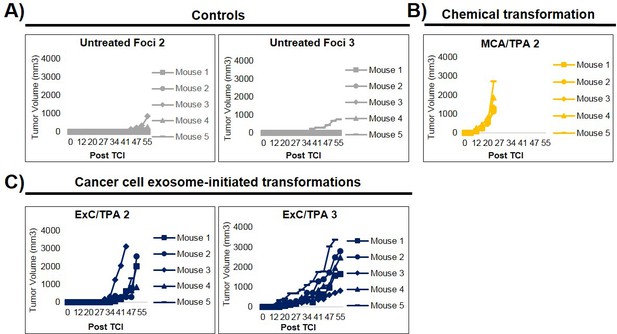
Additional in vivo studies of transformed cells.
(A) Results from additional foci tested from injection of background foci formed from untreated NIH/3T3 cells. Transformed cells were injected at a concentration of 1 × 106 cells. Results showed tumor growth in six out of 15 total mice (five tumors grew from ‘Untreated Foci 2’). (B) Additional independent focus tested from MCA/TPA transformed cells (n = 5). (C) Additional foci tested from Capan-2 exosome (ExC)/TPA transformed cells (n = 5).
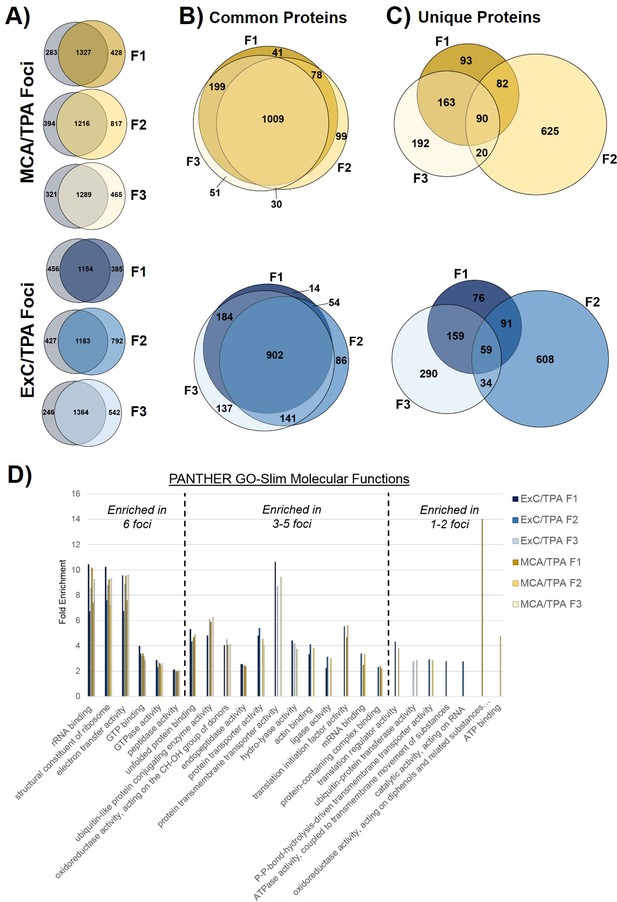
Proteomic profiling of transformed NIH/3T3 cells via mass spectrometry.
(A) Comparison of proteins found in transformed cells resulting from treatment with both an initiator and promoter visualized by Venn diagrams. Three separate foci (F1, F2, F3) from MCA/TPA transformed cells and Capan-2 exosome (ExC)/TPA transformed cells were compared to untreated NIH/3T3 cells (control, gray). Results from three biological replicates were combined for each sample. (B) Comparison of common (overlap) proteins found in each of the six transformed foci samples; common proteins identified in control. (C) Comparison of unique proteins found in each of the six transformed foci samples; unique proteins are absent from control. (D) Gene Ontology enrichment analysis of proteins found in all six foci using PANTHER 14.0. Slim molecular functions identified as overrepresented based on analysis of proteins found in samples.
-
Figure 4—source data 1
Relates to Figure 4.
Proteins identified by mass spectrometry analysis of transformed NIH/3T3 cells.
- https://doi.org/10.7554/eLife.40226.020
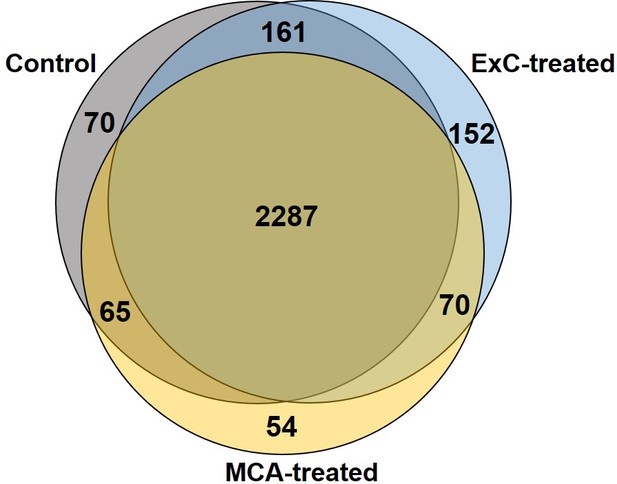
Proteomic profiling of initiated NIH/3T3 cells via mass spectrometry.
Comparison of proteins found in untreated NIH/3T3 cells (control) to cells treated with an initiator for 3 days (either MCA or Capan-2 (ExC) exosomes). For each condition, the results from three biological replicates were combined.
-
Figure 4—figure supplement 1—source data 1
Relates to Figure 4—figure supplement 1.
Proteins identified by mass spectrometry analysis of initiated NIH/3T3 cells.
- https://doi.org/10.7554/eLife.40226.019
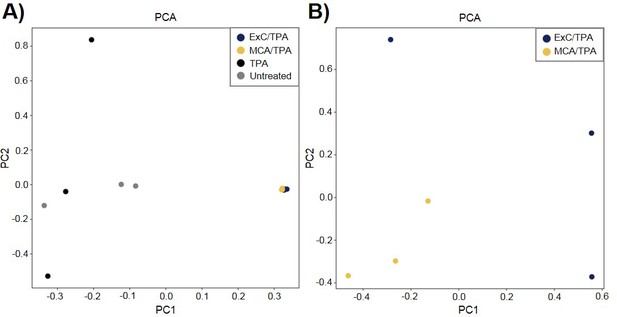
Principle component analysis (PCA) of transformed NIH/3T3 cells.
(A) PCA plot showing relationship between three MCA/TPA transformed foci, three Capan-2 exosome (ExC)/TPA transformed foci, three control foci from TPA-only treated NIH/3T3 cells, and three control foci from untreated NIH/3T3 cells. (B) PCA plot showing relationship between same three MCA/TPA transformed foci and Capan-2 exosome (ExC)/TPA transformed foci in the absence of control samples. Principle component analysis is based on comparison of exome-seq variant data using PLINK's identity-by-state (IBS) estimates.
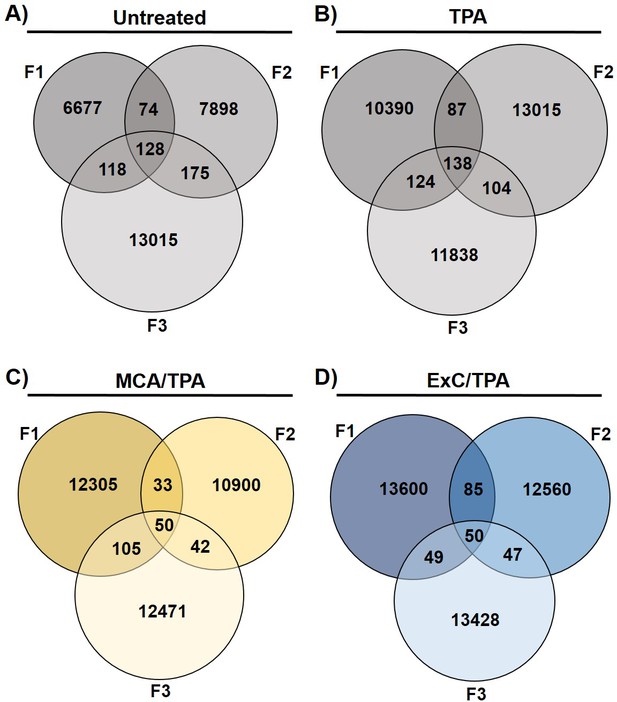
Variants found by Exome-sequencing analysis.
Total number of variants found in 12 samples sequenced by Exome-seq. Samples include transformed foci formed from four treatment conditions on NIH/3T3 cells: (A) untreated, (B) TPA-only treated, (C) MCA/TPA treated, and (D) Capan-2 exosome (ExC)/TPA treated.
-
Figure 5—figure supplement 1—source data 1
Relates to Figure 5—figure supplement 1.
Variants found in 190 oncogenes across all 12 samples analyzed in Figure 5.
- https://doi.org/10.7554/eLife.40226.023
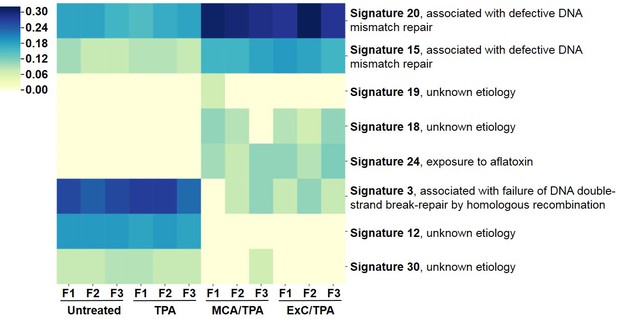
Mutational profiles of transformed NIH/3T3 cells.
The 12 samples sequenced via Exome-seq include transformed foci formed from four treatment conditions on NIH/3T3 cells: untreated, TPA-only treated, MCA/TPA treated, and Capan-2 exosome (ExC)/TPA treated. The top COSMIC mutational signatures associated with each sample were identified using MutaGene and clustered based on similarity to generate the heatmap shown. Color range corresponds to the contribution score of each mutational profile.
-
Figure 6—source data 1
Relates to Figure 6.
The top five mutational signatures found in each of the analyzed samples shown in Figure 6.
- https://doi.org/10.7554/eLife.40226.027
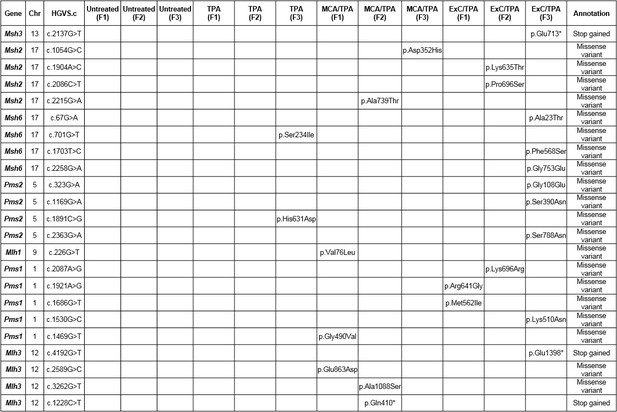
Non-synonymous variants found in mismatch repair associated genes (Msh2, Msh3, Msh6, Pms1, Pms2, Mlh1, Mlh3) across all 12 samples analyzed in Figure 5.
https://doi.org/10.7554/eLife.40226.025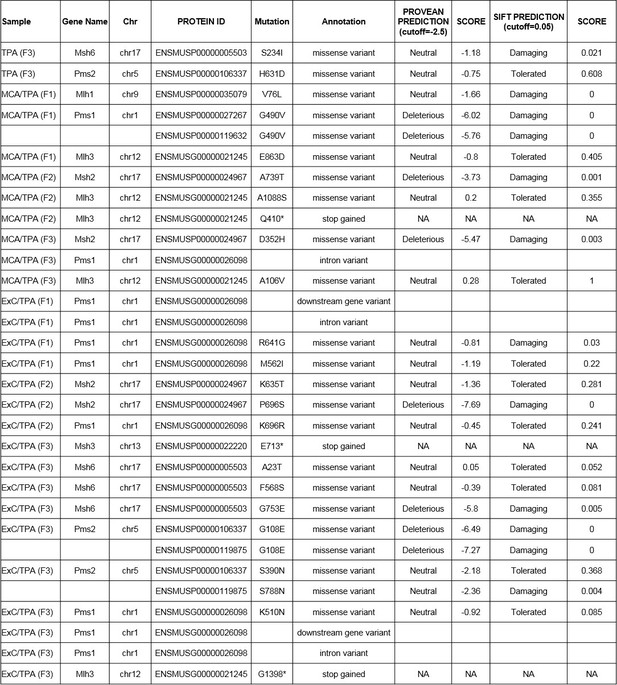
PROVEAN genome variant software was used to predict the potential impact of the identified missense variants on protein function in the mismatch repair associated genes.
https://doi.org/10.7554/eLife.40226.026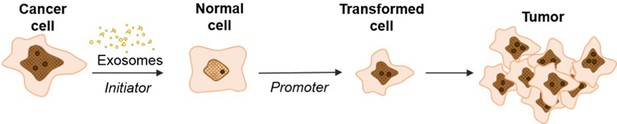
Schematic model of exosome mediated transformation.
Exosomes secreted by cancer cells are taken up by normal NIH/3T3 cells and have the capacity to act as an initiator by incorporating random changes into the recipient cell genome. These initiated cells, when exposed to a promoter, can be induced by further alterations to a transformed state that has the ability to grow into a malignant tumor.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical | 12-O-Tetradecanoylphorbol-13-acetate (TPA) | Cell Signaling Technology | 4174 | |
| Chemical | Methylcholanthrene (MCA) | Sigma-Aldrich | 213942–100 MG | |
| Chemical | Cadmium Chloride (CdCl2) | Sigma-Aldrich | 655198–5G | |
| Chemical | Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D2650−5 × 5 ML | |
| Cell line (Mus musculus) | NIH/3T3 | ATCC | RRID:CVCL_0594 | |
| Cell line (Homo-sapiens) | Dermal fibroblast (normal, Adult) | ATCC | PCS-201–012 | |
| Cell line (Homo-sapiens) | Capan-2 | ATCC | RRID:CVCL_0026 | |
| Cell line (Homo-sapiens) | PANC-1 | ATCC | RRID:CVCL_0480 | |
| Cell line (Homo-sapiens) | MIA PaCa-2 | ATCC | RRID:CVCL_0428 | |
| Cell line (Homo-sapiens) | BxPC-3 | ATCC | RRID:CVCL_0186 | |
| Cell line (Homo-sapiens) | HPDE (H6C7) | Kerafast | RRID:CVCL_0P38 | |
| Antibody | Anti-ALIX (3A9) (mouse monoclonal) | Abcam | Abcam, Cat#A2228, RRID:AB_10899268 | (1:500) |
| Antibody | α-actinin (H-2) (mouse monoclonal) | Santa Cruz Biotechnology | Santa Cruz Biotechnology, Cat#sc-17829, RRID:AB_626633 | (1:1000) |
| Antibody | Anti-β-actin (AC-74) (mouse monoclonal) | Sigma-Aldrich | Sigma-Aldrich, Cat#A2228, RRID:AB_476697 | (1:5000) |
| Antibody | Calnexin (C5C9) (rabbit monoclonal) | Cell Signaling Technology | Cell Signaling Technology Cat# 2679, RRID:AB_2228381 | (1:1000) |
| Antibody | CD63 (rabbit polyclonal) | Proteintech | Proteintech, Cat#25682–1-AP, RRID:AB_2783831 | (1:1000) |
| Antibody | HSP90α/β (F8) (mouse monoclonal) | Santa Cruz Biotechnology | Santa Cruz Biotechnology, Cat#sc-13119, RRID:AB_675659 | (1:1000) |
| Antibody | TSG101 (4A10) (mouse monoclonal) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# MA1-23296, RRID:AB_2208088 | (1:500) |
| Software, algorithm | GraphPad Prism 8 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Proteome Discoverer 2.1 | Thermo Scientific | Thermo Fisher Scientific, RRID:SCR_014477 | |
| Software, algorithm | FASTQC v0.11.5 | Babraham Bioinformatics | RRID:SCR_014583 | |
| Software, algorithm | Trim Galore | Babraham Bioinformatics | RRID:SCR_011847 | |
| Software, algorithm | Burrows-Wheeler Aligner (BWA) | Burrows-Wheeler Aligner | RRID:SCR_010910 | |
| Software, algorithm | Strelka2 | Illumina | RRID:SCR_005109 | |
| Software, algorithm | VCFtools | VCFtools | RRID:SCR_001235 | |
| Software, algorithm | SnpEff | SnpEff | RRID:SCR_005191 | |
| Software, algorithm | PLINK | Purcell et al., 2007 | RRID:SCR_001757 | |
| Software, algorithm | Python | Python Software Foundation | RRID:SCR_008394 | |
| Software, algorithm | MutaGene | MutaGene | RRID:SCR_016574 | |
| Software, algorithm | PROVEAN | J.Craig Venter Institute | RRID:SCR_002182 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40226.029





