Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism
Figures
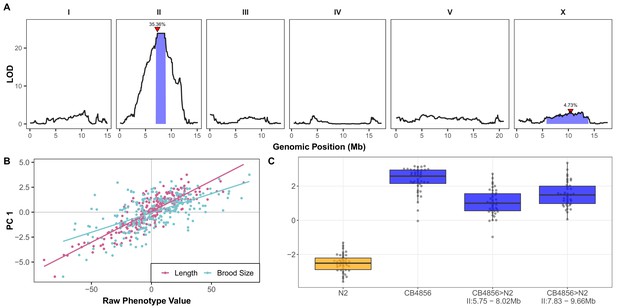
A large-effect QTL on the center of chromosome II explains differences in arsenic trioxide response between N2 and CB4856.
(A) Linkage mapping plots for the first principal component trait in the presence of 1000 µM arsenic trioxide is shown. The significance values (logarithm of odds, LOD, ratio) for 1454 markers between the N2 and CB4856 strains are on the y-axis, and the genomic position (Mb) separated by chromosome is plotted on the x-axis. The associated 1.5 LOD-drop confidence intervals are represented by blue boxes. The phenotypic variance explained by each QTL is shown above the peak QTL marker, which is marked by red triangles. (B) The correlation between brood size (blue; r2 = 0.38, p-value=1.65E-27) or animal length (pink; r2 = 0.74, p-value=3.16E-74) with the first principal component trait. Each dot represents an individual RIAIL’s phenotype, with the animal length and brood size phenotype values on the x-axis and the first principal component phenotype on the y-axis. (C) Tukey box plots of near-isogenic line (NIL) phenotype values for the first principal component trait in the presence of 1000 µM arsenic trioxide is shown. NIL genotypes are indicated below the plot as genomic ranges. The N2 trait is significantly different than the CB4856 and NIL traits (Tukey HSD p-value<1E-5).
-
Figure 1—source data 1
Arsenic dose-response loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
PCA was performed all arsenic concentrations together in order to look at the relative PC value for each concentration.
- https://doi.org/10.7554/eLife.40260.025
-
Figure 1—source data 2
RIAIL phenotype data used for linkage mapping.
(Used to generate Figure 1B and Figure 1—figure supplement 4).
- https://doi.org/10.7554/eLife.40260.026
-
Figure 1—source data 3
Results from linkage mapping experiment.
(Used to generate Figure 1A, Figure 1—figure supplement 5, and Figure 1—figure supplement 7).
- https://doi.org/10.7554/eLife.40260.027
-
Figure 1—source data 4
Phenotypes of NILs and CRISPR allele replacement strains in the presence of 1000 µM arsenic trioxide after correcting for strain differences in control conditions.
(Used to generate Figure 1C, Figure 3, Figure 1—figure supplement 8 and Figure 3—figure supplement 1).
- https://doi.org/10.7554/eLife.40260.028
-
Figure 1—source data 5
NIL genotypes generated from whole-genome sequencing.
- https://doi.org/10.7554/eLife.40260.029
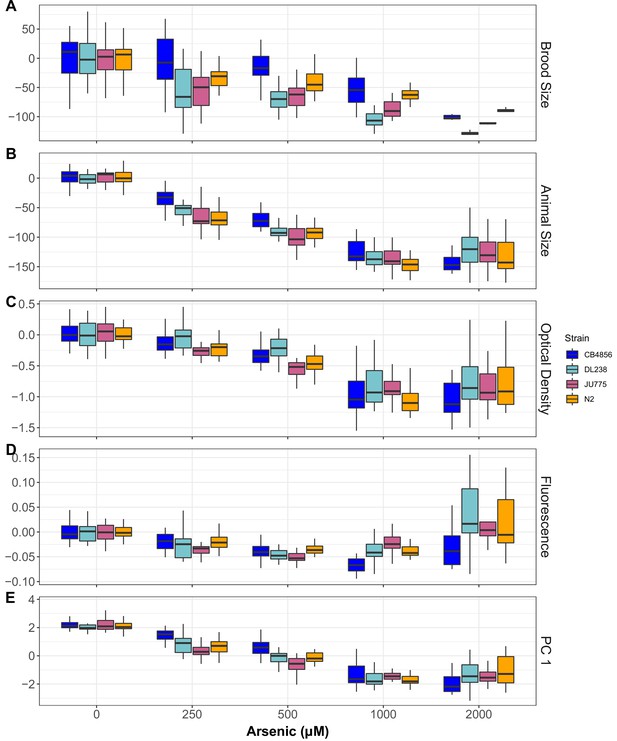
Arsenic trioxide dose response of four diverged C. elegans strains.
Arsenic trioxide concentration in µM is plotted on the x-axis and the (A) normalized brood size, (B) mean progeny length, (C) mean normalized optical density, (D) mean normalized yellow fluorescence, or (E) the first principal component are plotted on the y-axes. For panels A-D, the y-axis values represent individual phenotypic measurements subtracted from the mean value in 0 µM arsenic trioxide. At least 15 replicates for each strain and condition are represented by Tukey box plots. Box plots are colored by strain (CB4856:blue, DL238:teal, JU775:pink, and N2:orange).
-
Figure 1—figure supplement 1—source data 1
Arsenic dose-response trait data for four strains in 0, 250, 500, 1000, or 2000 µM arsenic, labeled as water, arsenic1, arsenic2, arsenic3, arsenic4 in the Condition column.
(Used to generate Figure 1—figure supplement 1A-B).
- https://doi.org/10.7554/eLife.40260.004
-
Figure 1—figure supplement 1—source data 2
Arsenic dose-response principal component (PC) eigenvectors for the PCs that explain up to 90% of the total variance in the data set.
PCA was performed all arsenic concentrations together in order to look at the relative PC value for each concentration. Condition names correspond to those in Supplementary file 1. (Used to generate Figure 1—figure supplement 1C).
- https://doi.org/10.7554/eLife.40260.005
-
Figure 1—figure supplement 1—source data 3
Arsenic dose-response trait correlations where each row corresponds to the Pearson correlation coefficient for two traits.
Condition names correspond to those in Supplementary file 1. (Used to generate Figure 1—figure supplement 1A-D).
- https://doi.org/10.7554/eLife.40260.006
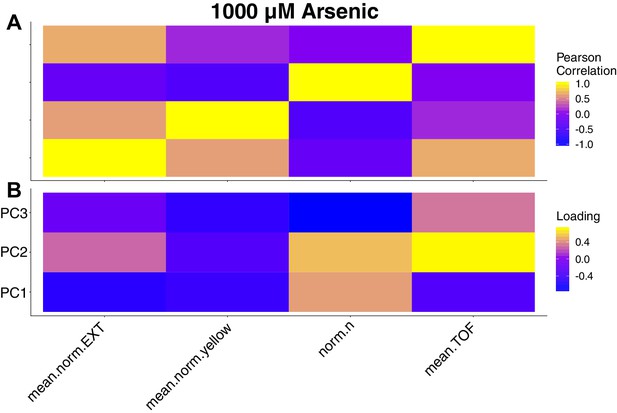
Trait correlations and principal component loadings of arsenic trioxide dose response.
(A) The trait Pearson’s correlation coefficient of the assay- and control-regressed measured traits from the dose response experiment with 1000 µM arsenic trioxide is shown. (B) The contribution of each measured trait to the principal components that explain 90% of the total variance is shown. For each plot, the tile color corresponds to the value, where yellow colors represent higher values.
-
Figure 1—figure supplement 2—source data 1
Arsenic dose-response loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
Condition names correspond to those in Supplementary file 1. (Used to generate Figure 1—figure supplement 2B).
- https://doi.org/10.7554/eLife.40260.008
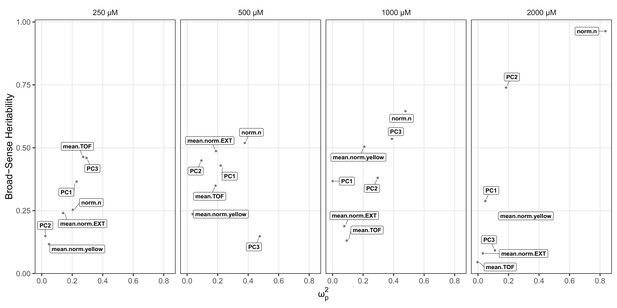
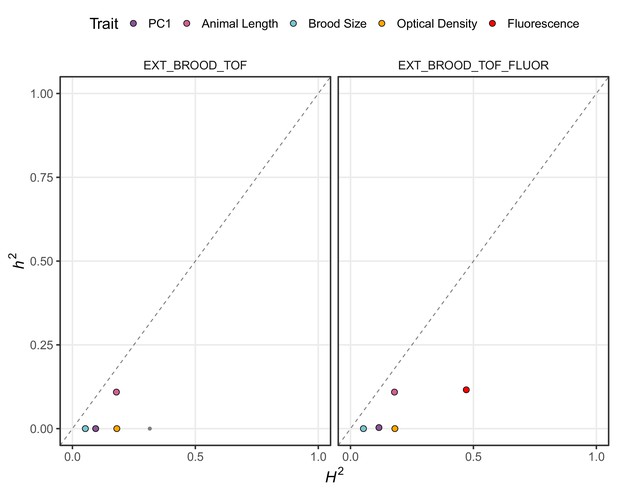
Effect size and broad-sense heritability estimates for the arsenic trioxide dose response.
Each panel corresponds to a concentration of arsenic trioxide concentration, which is indicated above the plot panel. The x-axis represents the partial omega squared () effect-size estimate, and the y-axis represent the broad-sense heritability estimate (H2). Each dot represents a different BIOSORT-measured or principal component trait.
-
Figure 1—figure supplement 3—source data 1
Broad-sense heritability estimates from arsenic dose response data.
Estimates were calculated using the lmer function of the lme4 R package by fitting the mixed effect model equation lmer(value ~ (1|strain)), where value is the trait value and strain is the strain name. (Used to generate Figure 1—figure supplement 3).
- https://doi.org/10.7554/eLife.40260.010
-
Figure 1—figure supplement 3—source data 2
Arsenic dose-response estimates of effect sizes estimated by fitting the linear model: trait value ~strain.
(Used to generate Figure 1—figure supplement 3).
- https://doi.org/10.7554/eLife.40260.011
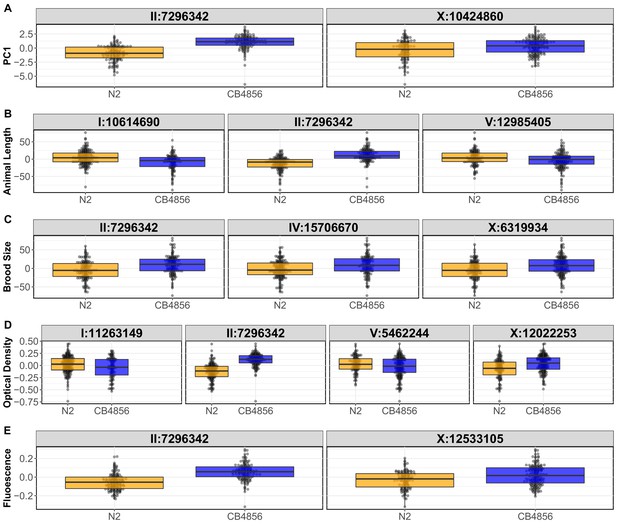
RIAIL phenotypes from the linkage mapping experiment.
Tukey box plots of the first principal component are shown: (A), assay- and control-regressed mean animal length (B), brood sizes (C), optical density (D), and mean normalized yellow fluorescence (E) of the N2 and CB4856 RIAIL panel after exposure to arsenic trioxide. Each dot corresponds to the phenotype for a single RIAIL. The RIAILs are separated by the N2 (orange) or CB4856 (blue) genotype at each QTL detected by linkage mapping.
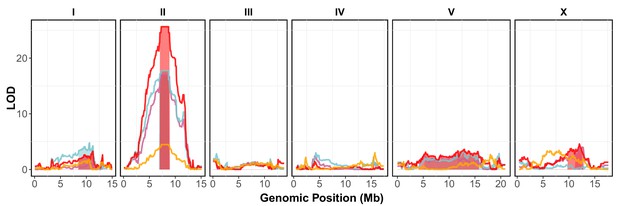
Linkage mapping results for brood size, animal length, and the first principal component.
Linkage mapping plots for regressed brood size (orange), mean normalized yellow fluorescence (pink), mean animal length (blue) and mean normalized optical density (red) in the presence of 1000 µM arsenic trioxide are shown. The significance values (logarithm of odds (LOD) ratio) for 1454 markers between the N2 and CB4856 strains are on the y-axis, and the genomic position (Mb) separated by chromosome is plotted on the x-axis. The associated 1.5 LOD-drop confidence intervals are represented by colored boxes below significant QTL.
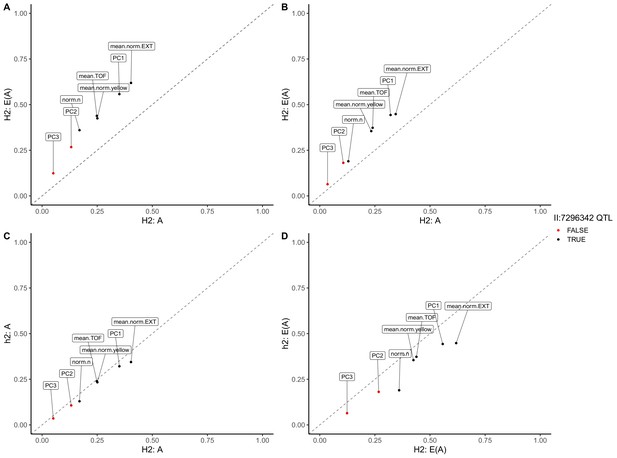
Genomic-heritability estimates of linkage mapping traits.
The genomic broad (H2)- and narrow (h2)-sense heritability estimates calculated using the expectation (E(A)) of the realized relatedness matrix or the realized relatedness matrix (A) are shown. Each dot represents a measured or principal component trait. Dots are colored black if that trait mapped to the center of chromosome II and red if no QTL was detected on the center of chromosome II.
-
Figure 1—figure supplement 6—source data 1
Genomic heritability estimates from RIAIL phenotype data.
(Used to generate Figure 1—figure supplement 6).
- https://doi.org/10.7554/eLife.40260.015
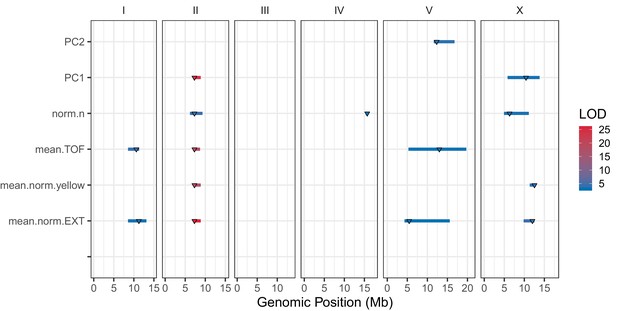
Linkage mapping QTL summary.
All QTL identified by linkage mapping are shown. Traits are labeled on the y-axis and the genomic position in Mb is plotted on the x-axis. Only PCs that explain greater than 90% of the variance are shown. Triangles represent the peak QTL position and bars represent the associated 1.5-LOD drop QTL confidence interval. Triangles and bars are colored based on the LOD score, where red colors correspond to higher LOD values.
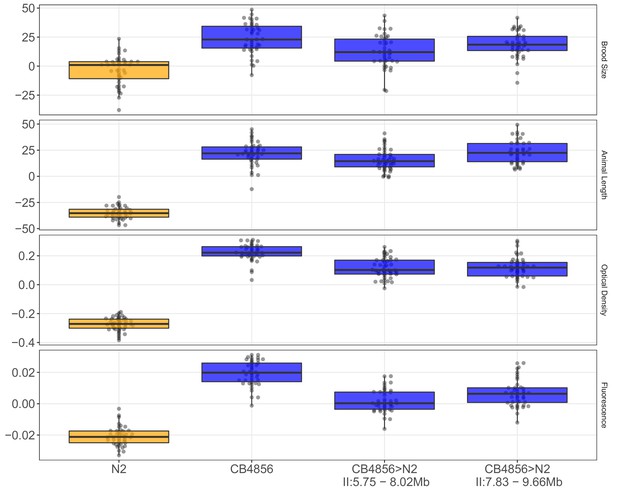
NIL recapitulation of chromosome II QTL.
Tukey box plots of near-isogenic line (NIL) phenotype values for, from top to bottom: the brood size, mean animal length, mean normalized optical density, and mean normalized yellow fluorescence traits in the presence of 1000 µM arsenic trioxide are shown. NIL genotypes are indicated below the plot as genomic ranges. The N2 phenotypes are significantly different from all other strains (Brood size: Tukey HSD p-value=2E-7; Animal Length: Tukey HSD p-value<1E-7; Optical Density: Tukey HSD p-value<1E-7; Fluorescence: Tukey HSD p-value<1E-7).
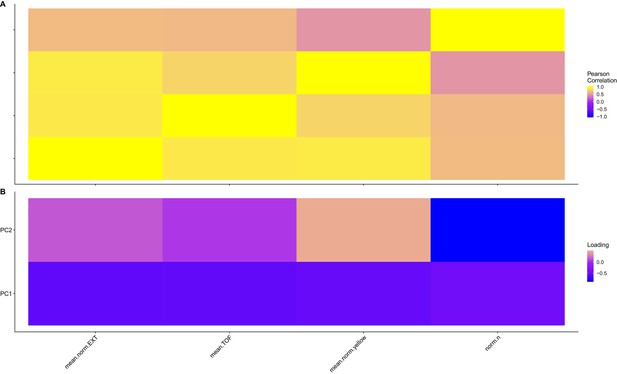
Trait correlations and principal component loadings of NIL and allele-replacement recapitulation experiment.
(A) The Pearson’s correlation coefficients of the assay- and control-regressed measured traits are shown. (B) The contribution of each measured trait to the principal components that explain 90% of the total variance in the NIL and allele replacement-recapitulation experiment, which was performed at 1000 µM, is shown. For each plot, the tile color represents the value, where yellow colors represent higher values.
-
Figure 1—figure supplement 9—source data 1
NIL and CRISPR allele replacement trait correlations.
(Used to generate Figure 1—figure supplement 9A).
- https://doi.org/10.7554/eLife.40260.019
-
Figure 1—figure supplement 9—source data 2
NIL and CRISPR allele replacement trait loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
(Used to generate Figure 1—figure supplement 9B).
- https://doi.org/10.7554/eLife.40260.020
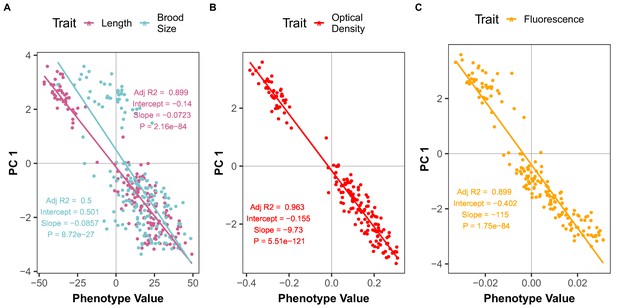
Brood size and animal length are correlated with the first principal component for the NIL recapitulation experiment.
The correlation between brood size (blue) or animal length (pink) (A), mean normalized optical density (B) or mean normalized yellow fluorescence (C) traits with the first principal component (PC1) trait for the NIL recapitulation experiment are shown. Each dot represents an individual NIL or parental strain replicate phenotype with the animal length and brood size phenotype values on the x-axis and the first principal component (PC1) phenotype on the y-axis.
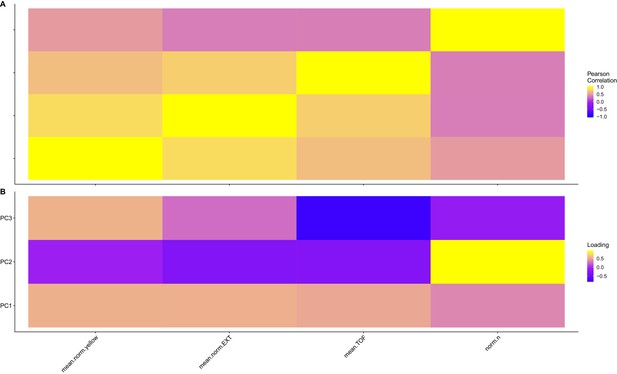
Trait correlations and principal component loadings of linkage mapping experiment.
(A) The trait Pearson’s correlation coefficient of the assay- and control-regressed measured traits is shown. (B) The contribution of each measured trait to the principal components that explain 90% of the total variance in the linkage mapping experiment, which was performed at 1000 µM, is shown. For each plot, the tile color represents the value, where yellow colors represent higher values.
-
Figure 1—figure supplement 11—source data 1
RIAIL trait correlations where each row corresponds to the Pearson correlation coefficient for two traits.
(Used to generate Figure 1—figure supplement 11A).
- https://doi.org/10.7554/eLife.40260.023
-
Figure 1—figure supplement 11—source data 2
RIAIL loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
(Used to generate Figure 1—figure supplement 11B).
- https://doi.org/10.7554/eLife.40260.024
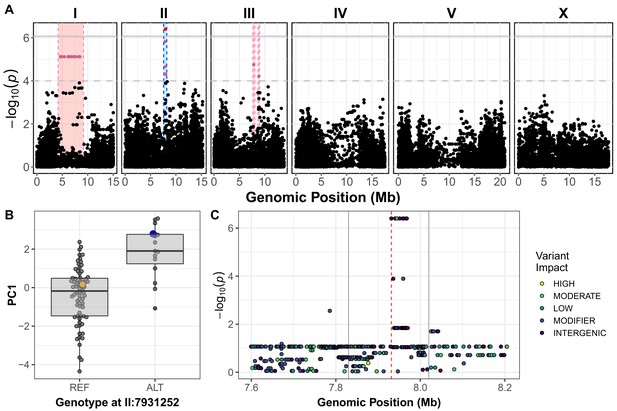
Variation in C. elegans wild isolates responses to arsenic trioxide maps to the center of chromosome II.
(A) A manhattan plot for the first principal component in the presence of 1000 µM arsenic trioxide is shown. Each dot represents an SNV that is present in at least 5% of the assayed wild population. The genomic position in Mb, separated by chromosome, is plotted on the x-axis and the -log10(p) for each SNV is plotted on the y-axis. SNVs are colored red if they pass the genome-wide Bonferroni-corrected significance (BF) threshold, which is denoted by the gray horizontal line. SNVs are colored pink if they pass the genome-wide eigen-decomposition significance (ED) threshold, which is denoted by the dotted gray horizontal line. The genomic region of interests surrounding the QTL that pass the BF and ED thresholds are represented by cyan and pink rectangles, respectively. (B) Tukey box plots of phenotypes used for association mapping in (A) are shown. Each dot corresponds to the phenotype of an individual strain, which is plotted on the y-axis. Strains are grouped by their genotype at the peak QTL position (red SNV from panel A, ChrII:7,931,252), where REF corresponds to the allele from the reference N2 strain. The N2 (orange) and CB4856 (blue) strains are highlighted. (C) Fine mapping of the chromosome II region of interest (cyan region from panel A, 7.60–8.21 Mb) is shown. Each dot represents an SNV present in the CB4856 strain. The association between the SNV and first principal component is shown on the y-axis and the genomic position of the SNV is shown on the x-axis. Dots are colored by their SnpEff predicted effect.
-
Figure 2—source data 1
Fine-mapping results for PC1.
(Used to generate Figure 2C and Figure 2—figure supplement 2).
- https://doi.org/10.7554/eLife.40260.040
-
Figure 2—source data 2
All wild-isolate traits used for genome-wide association mapping.
- https://doi.org/10.7554/eLife.40260.041
-
Figure 2—source data 3
Genotype matrix used for genome-wide mapping.
- https://doi.org/10.7554/eLife.40260.044
-
Figure 2—source data 4
GWA mapping results for PC1.
(Used to generate Figure 2A–B).
- https://doi.org/10.7554/eLife.40260.045
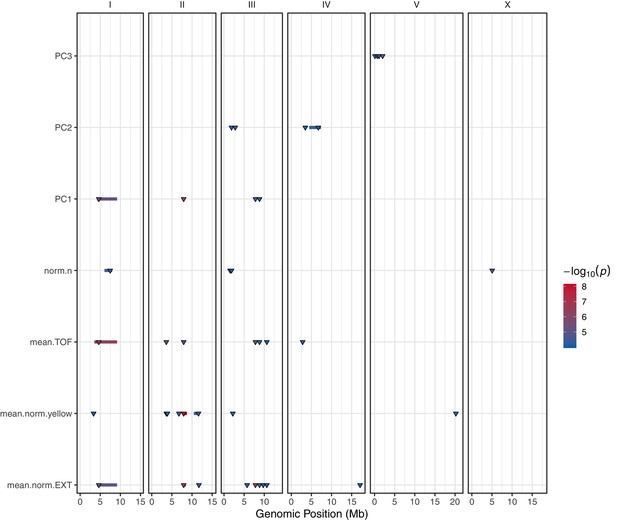
GWA mapping QTL summary.
All QTL identified by GWA mapping above the eigen significance threshold are shown. Traits are labeled on the y-axis and the genomic position in Mb is plotted on the x-axis. Only PCs that explain greater than 90% of the variance are shown. Triangles represent the peak QTL position, and bars represent the associated QTL region of interest. Triangles and bars are colored based on the significance value where red colors correspond to higher significance values.
-
Figure 2—figure supplement 1—source data 1
All QTL identified by GWA mapping (Used to generate Figure 2—figure supplement 1).
- https://doi.org/10.7554/eLife.40260.032
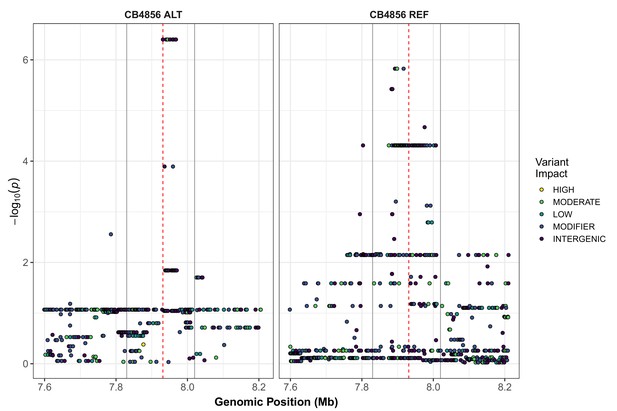
Fine-mapping of the chromosome II QTL identified by GWA mapping.
Fine mapping of the chromosome II region of interest (cyan region from Figure 1A, 7.71–8.18 Mb) is shown. Each dot represents an SNV present in the phenotyped population. SNVs present in the CB4856 strain are shown in the left panel and SNVs present in other phenotyped strains, but REF in CB4856, are shown in the right panel. The association between the SNV and first principal component (PC1) is shown on the y-axis, and the genomic position of the SNV is shown on the x-axis. Dots are colored by their SnpEff predicted effect.
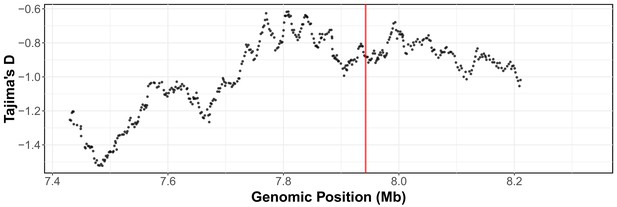
Tajima’s D across the arsenic trioxide QTL confidence interval.
Divergence, as measured by Tajima's D, is shown across the arsenic trioxide QTL confidence interval (II:7,430,000–8,330,000). The whole-genome SNV data set (Cook et al., 2016; Cook et al., 2017) was used for Tajima’s D calculations. Window size for the calculations was 500 SNVs with a 10 SNV sliding window size. The vertical red line marks the position of the dbt-1 locus.
-
Figure 2—figure supplement 3—source data 1
Isolation locations of strains used in GWA mapping.
(Used to generate Figure 2—figure supplement 3).
- https://doi.org/10.7554/eLife.40260.035
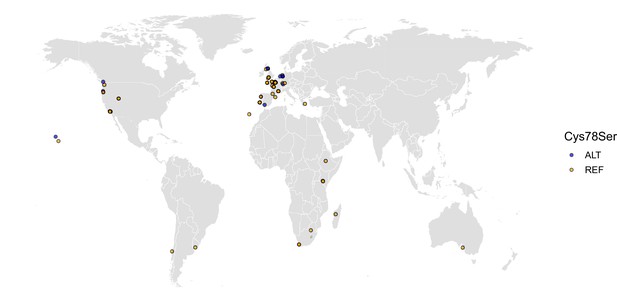
The worldwide distribution of the DBT-1(C78S) allele.
Cysteine (REF) is shown in orange, and serine (ALT) is shown in blue. Latitude and longitude coordinates of sampling locations were used to plot individual strains on the map.
Genomic-heritability estimates of GWA mapping traits.
The genomic broad (H2)- and narrow (h2)-sense heritability estimates calculated using the realized relatedness matrix are shown. Each dot represents a measured or principal component trait. The five traits discussed throughout the manuscript are highlighted (purple: first principal component, pink: mean animal length, blue: brood size, orange; mean normalized optical density, and red: mean normalized yellow fluorescence). The four input traits for PCA in the left panel are mean normalized optical density, brood size, and mean animal length. For the right panel, mean normalized yellow fluorescence was also added.
-
Figure 2—figure supplement 5—source data 1
Genomic heritability estimates of wild isolate traits.
(Used to generate Figure 2—figure supplement 5).
- https://doi.org/10.7554/eLife.40260.038
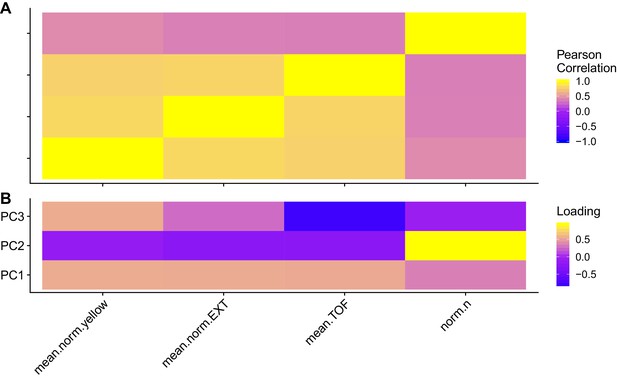
Trait correlations and principal component loadings of GWA mapping experiment.
(A) The Pearson’s correlation coefficients of the assay- and control-regressed for measured traits are shown. (B) The contribution of each measured traits to the principal components that explain 90% of the total variance in the GWA mapping experiment, which was performed at 1000 µM, is shown. For each plot, the tile color represents the value, where yellow colors represent higher values.
-
Figure 2—figure supplement 6—source data 1
Wild isolate trait loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
(Used to generate Figure 2—figure supplement 6B).
- https://doi.org/10.7554/eLife.40260.042
-
Figure 2—figure supplement 6—source data 2
Wild isolate trait correlations.
(Used to generate Figure 2—figure supplement 6A).
- https://doi.org/10.7554/eLife.40260.043
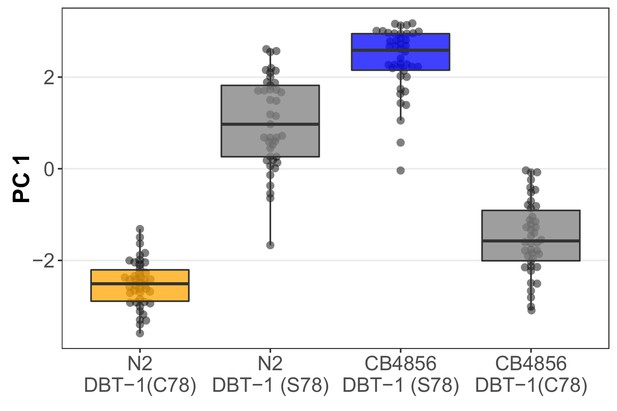
The DBT-1(C78S) variant contributes to arsenic trioxide responses.
Tukey box plots of the first principal component generated by PCA on allele-replacement strainphenotypes measured by the COPAS BIOSORT 1000 μM arsenic trioxide exposure are shown (N2,orange; CB4856, blue; allele replacement strains, gray). Labels correspond to the genetic backgroundand the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). All pair-wise comparisons are significantly different (Tukey HSD, p-value < 1E-7).
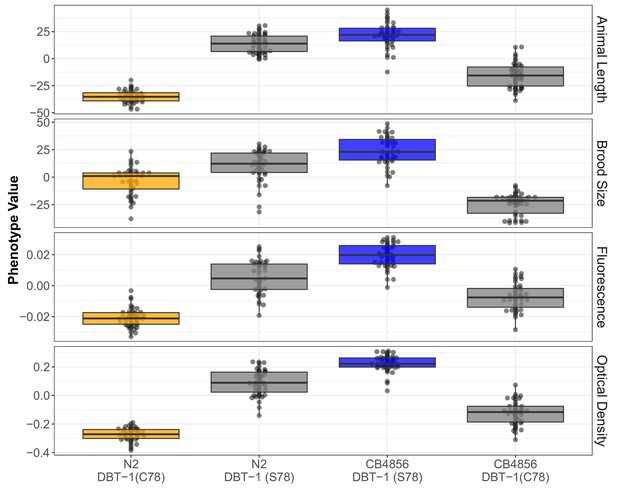
The DBT-1 C78S variant underlies arsenic trioxide sensitivity in C. elegans.
From top to bottom: Tukey box plots of residual mean animal length, brood size, mean normalized yellow fluorescence, and mean normalized optical density after exposure to 1000 µM arsenic trioxide are shown (N2, orange; CB4856, blue; allele replacement strains, gray). Labels correspond to the genetic background and the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). Every pairwise strain comparison was significant (Brood size: Tukey’s HSD p-value<1.55E-5; Animal length: p-value<2E-73; Fluorescence: p-value<1E-7; Optical Density: p-value<1E-7).
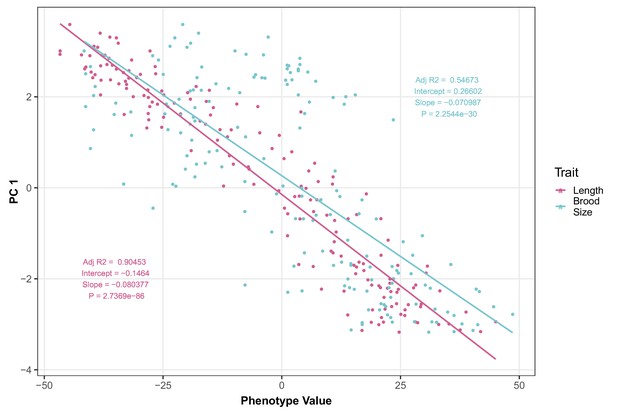
Brood size and animal length are correlated with the first principal component for the allele- replacement recapitulation experiment.
The correlations between brood size (blue) or animal length (pink) with the first principal component trait for the allele- replacement recapitulation experiment are shown. Each dot represents an individual allele-replacement or parental strain replicate phenotype, with the animal length and brood size phenotype values on the x-axis and the first principal component (PC1) phenotype on the y-axis.
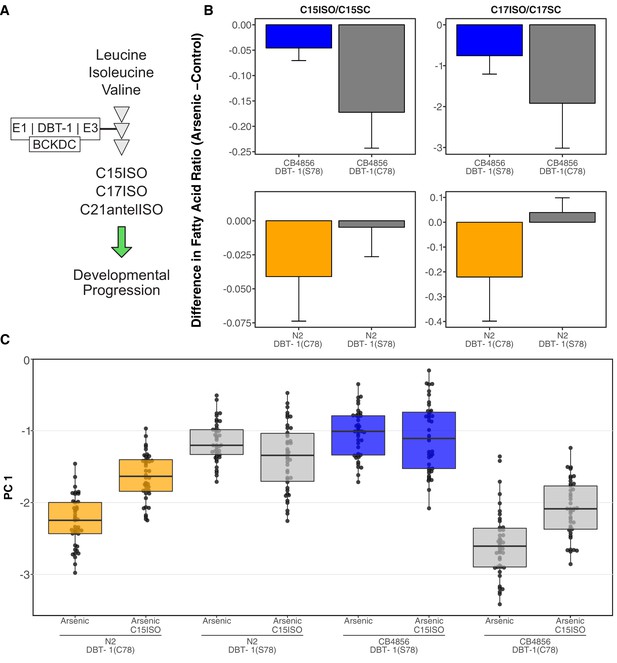
Differential production of mmBCFA underlies DBT-1(C78)-mediated sensitivity to arsenic trioxide.
(A) A simplified model of BCAA catabolism in C. elegans. The BCKDH complex, which consists of DBT-1, catalyzes the irreversible oxidative decarboxylation of branched-chain ketoacids. The products of thesebreakdown can then serve as building blocks for the mmBCFA that are required for developmental progression. (B) The difference in the C15ISO/C15SC (left panel) or C17ISO/C17SC (right panel) ratios between 100 μM arsenic trioxide and control conditions is plotted on the y-axis for three independent replicates of the CB4856 and CB4856 allele replacement strains and six independent replicates of the N2and N2 allele replacement strains. The difference between the C15 ratio for the CB4856-CB4856 allele replacement comparison is significant (Tukey HSD p-value = 0.0427733), but the difference between the C17 ratios for these two strains is not (Tukey HSD p-value = 0.164721). The difference between the C15and C17 ratios for the N2-N2 allele replacement comparisons are both significant (C15: Tukey HSD p-value = 0.0358; C17: Tukey HSD p-value = 0.003747). (C) Tukey box plots median animal length after arsenic trioxide or arsenic trioxide and 0.64 μM C15ISO exposure are shown (N2, orange; CB4856, blue; allele replacement strains, gray). Labels correspond to the genetic background and the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). Every pair-wise strain comparison is significant except for the N2 DBT-1(S78) - CB4856 comparisons (Tukey’s HSD p-value < 1.43E-6).
-
Figure 4—source data 1
Metabolite measurements for the CB4856 and CB4856 allele replacement strains (Used for Figure 4B and Figure 4—figure supplement 1–3).
- https://doi.org/10.7554/eLife.40260.062
-
Figure 4—source data 2
Processed metabolite measurements for the CB4856 and CB4856 allele replacement strains (Used for Figure 4B and Figure 4—figure supplement 1–3).
- https://doi.org/10.7554/eLife.40260.063
-
Figure 4—source data 3
Metabolite measurements for the N2 and N2 allele replacement strains (Used for Figure 4B and Figure 4—figure supplement 1–3).
- https://doi.org/10.7554/eLife.40260.064
-
Figure 4—source data 4
Processed phenotype data for the C15ISO rescue experiment (Used to generate Figure 4C).
- https://doi.org/10.7554/eLife.40260.065
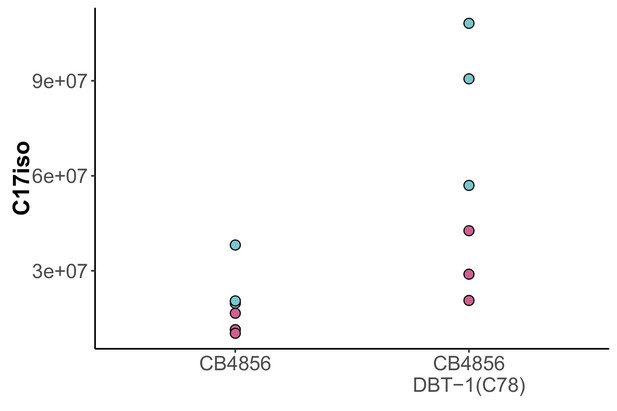
Raw abundance of C17ISO for CB4856 and CB4856 allele replacement.
The raw abundance of C17ISO is plotted on the y-axis for three independent replicates of the CB4856 and CB4856 allele replacement strains exposed to control (teal) or 100 µM arsenic trioxide (pink) conditions. The difference between CB4856 replacement mock and arsenic conditions was significant (Tukey HSD p-value=0.029), but the difference between CB4856 mock and arsenic conditions was not significant (Tukey HSD p-value=0.10).
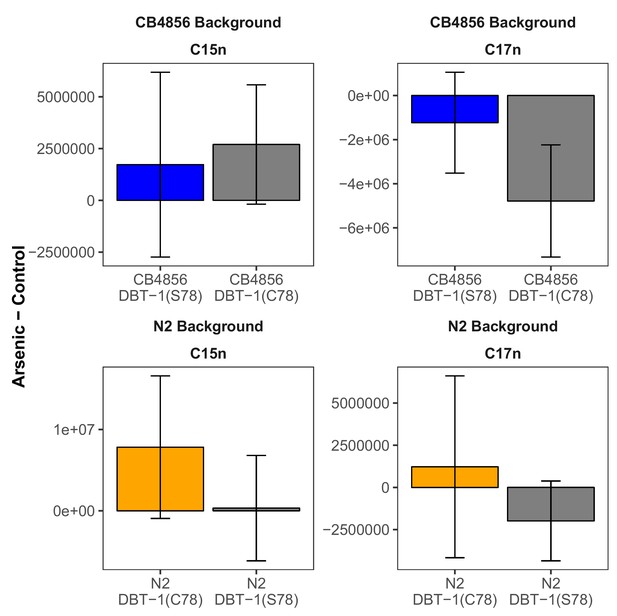
Straight-chain fatty acids are not affected by arsenic trioxide.
The difference in raw C15SC (left panel) or C17SC (right panel) abundances between 100 µM arsenic trioxide and control conditions is plotted on the y-axis for three independent replicates of the CB4856 and CB4856 allele replacement strains and six independent replicates of the N2 and N2 allele replacement strains. We found no significant differences when comparing the abundances between parental and allele-replacement strains.
-
Figure 4—figure supplement 2—source data 1
C15ISO rescue trait loadings of principal components (PCs) for the PCs that explain up to 90% of the total variance in the trait data.
(Used to generate Figure 4—figure supplement 2B).
- https://doi.org/10.7554/eLife.40260.052
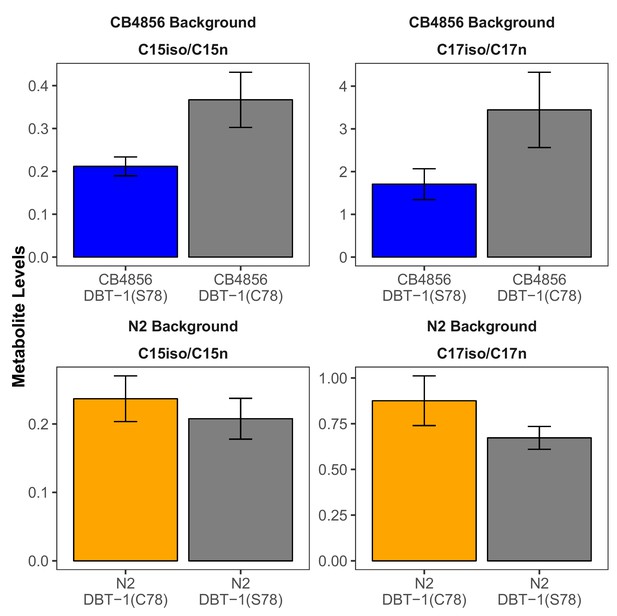
C15ISO and C17ISO to strait-chain ratios in control conditions.
The C15ISO/C15SC (left panel) or C17ISO/C17SC (right panel) ratios in control conditions are plotted on the y-axis for three independent replicates of the CB4856 and CB4856 allele replacement strains and six independent replicates of the N2 and N2 allele replacement strains. The C15 and C17 ratios for the CB4856-CB4856 allele replacement comparison were significant (C15: Tukey HSD p-value=0.0168749; C17: Tukey HSD p-value=0.0342525). The difference between the C17 ratio for the N2-N2 allele replacement comparison was significant (Tukey HSD p-value=0.0044667), but the difference in the C15 ratio was not significant (Tukey HSD p-value=0.1239674).
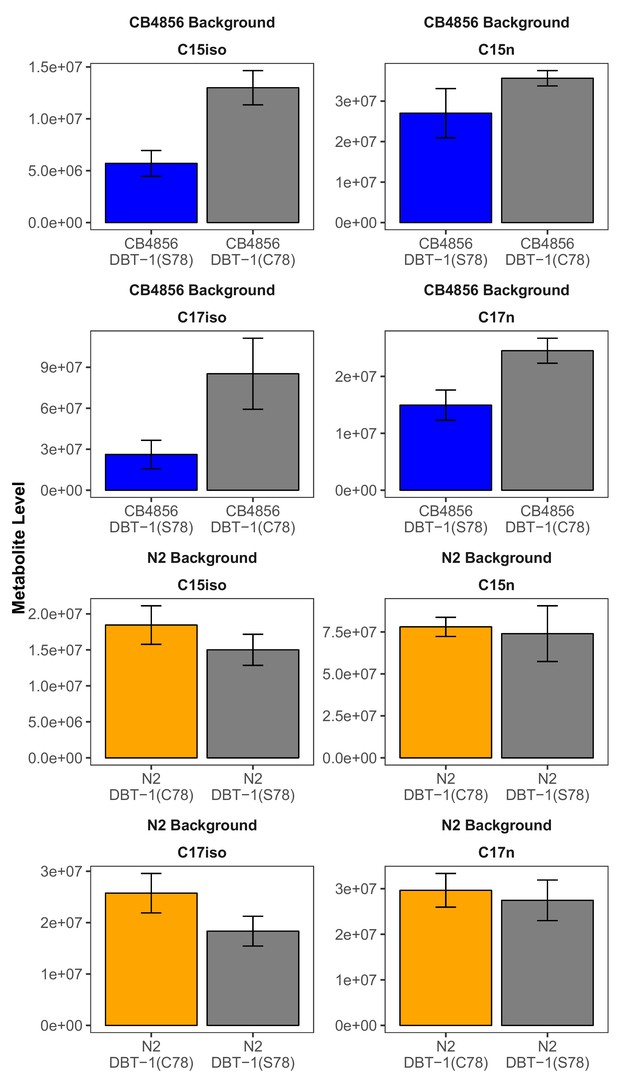
Strains with the DBT-1(C78) allele produce more branched chain fatty acids in the L1 larval stage in control conditions.
Branched chain (left panel) and straight chain (right panel) fatty acid measurements in L1 animals arerepresented on the y-axis. We found significant differences in abundances when comparing all parentaland allele-replacement strains for C15ISO and C17ISO chain fatty acids (CB4856-C15ISO DBT-1(C78):Tukey HSD p-value = 0.0036201, n=3; N2-C15ISO DBT-1(C78): Tukey HSD p-value = 0.0265059, n=6;CB4856-C17ISO DBT-1(C78): Tukey HSD p-value = 0.0086572, n=3; N2-C17ISO DBT-1(C78): TukeyHSD p-value = 0.0022501, n=6). Conversely, we observed no significant differences in straight chain fattyproduction except for C17n production in the CB4856 background (CB4856-C15n DBT-1(C78): TukeyHSD p-value = 0.0787388, n=3; N2-C15n DBT-1(C78): Tukey HSD p-value = 0.5817993, n=6; CB4856-C17n DBT-1(C78): Tukey HSD p-value = 0.0086572, n=3; N2-C17n DBT-1(C78): Tukey HSD p-value =0.35827, n=6).
-
Figure 4—figure supplement 4—source data 1
Metabolite measurements for N2, CB4856, and both allele-replacement strains at the L4 larval stage.
(Used to generate Figure 4—figure supplement 4).
- https://doi.org/10.7554/eLife.40260.055
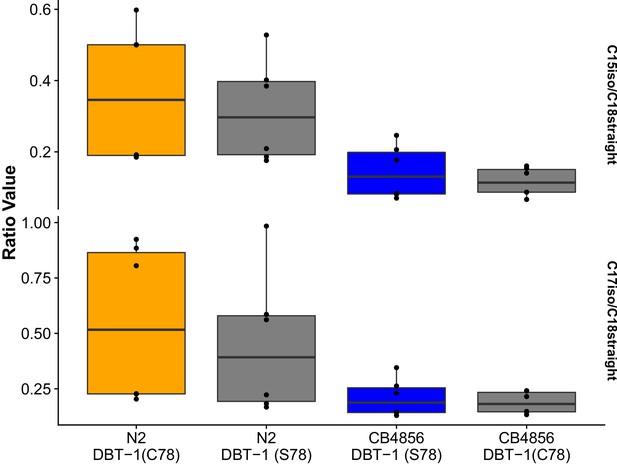
Young adult C15ISO and C17ISO to strait-chain ratios in control conditions.
The C15ISO/C18SC (top panel) or C17ISO/C18SC(bottom panel) ratios of young adult animals in control conditions are plotted on the y-axis for six independent replicates for all strains. We found no significant differences when comparing the abundances between parental and allele-replacement strains.
-
Figure 4—figure supplement 5—source data 1
Trait correlations for the for the C15ISO rescue experiment (Used to generate Figure 4—figure supplement 5).
- https://doi.org/10.7554/eLife.40260.057
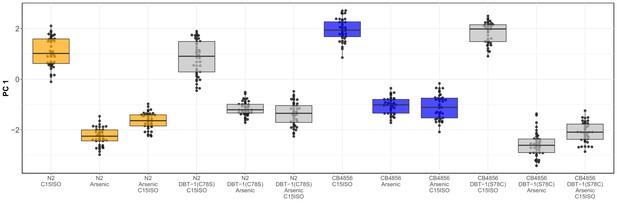
Complete results from C15iso rescue experiment.Tukey box plots for the PC1 trait after 0.64 µM C15ISO, arsenic trioxide, or arsenic trioxide and 0.64 µM C15ISO exposure are shown (N2, orange; CB4856, blue; allele replacement strains, gray).
Labels correspond to the genetic background and the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). Every pairwise strain comparison was significant except for the N2 DBT-1(S78) - CB4856 comparisons (Tukey’s HSD p-value<1.43E-6).
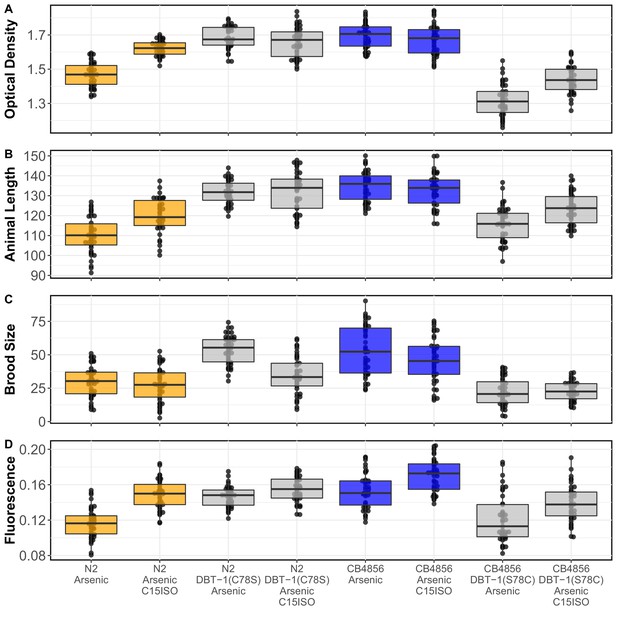
BIOSORT-quantified traits for C15ISO rescue experiment Tukey box plots for the mean normalized optical density.
(A), mean animal length (B), brood size (C), and mean normalized yellow fluorescence (D) traits after 0.64 µM C15ISO, arsenic trioxide, or arsenic trioxide and 0.64 µM C15ISO exposure are shown (N2, orange; CB4856, blue; allele replacement strains, gray). Labels correspond to the genetic background and the corresponding residue at position 78 of DBT-1 (C for cysteine, S for serine). For all traits, except brood size, the comparison between arsenic and arsenic with C15ISO for strains with the DBT-1(C78) allele was significant (Tukey’s HSD p-value<2E-3).
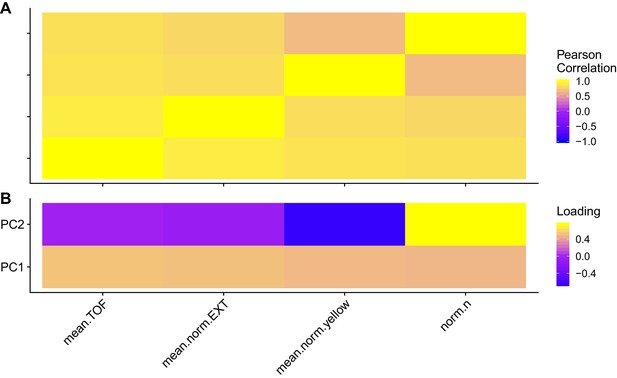
Trait correlations and principal component loadings of C15ISO rescue experiment.
(A) The Pearson’s correlation coefficients of the assay- and control-regressed measured traits are shown. (B) The contribution of each measured trait to the principal components that explain 90% of the total variance in the GWA mapping experiment, which was performed at 1000 µM, is shown. For each plot, the tile colors represent the value, where yellow colors represent higher values.
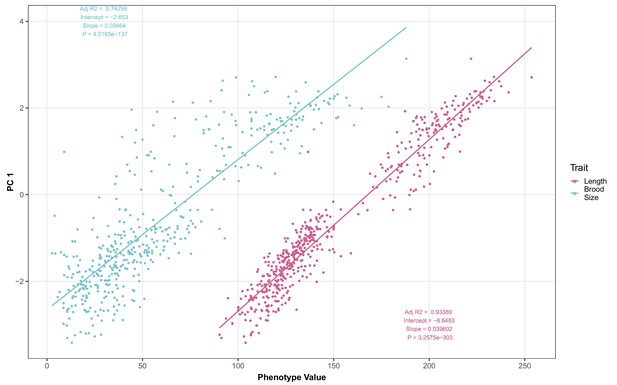
Brood size and animal length are correlated with the first principal component for the C15ISO rescue experiment.
The correlations between brood size (blue) or animal length (pink) with the first principal component trait for the C15ISO rescue experiment are shown. Each dot represents an individual allele-replacement or parental strain replicate phenotype, with the animal length and brood size phenotype values on the x-axis and the first principal component phenotype on the y-axis.
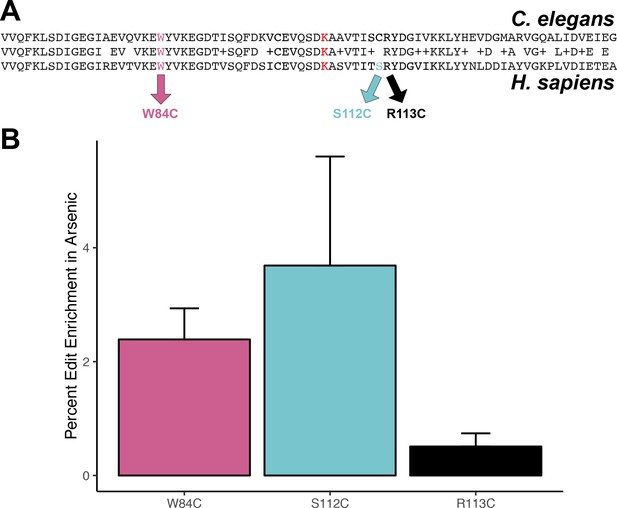
Protective effect of cysteine residues in human DBT1.
(A) Alignment of C. elegans DBT-1 and H. sapiens DBT1. The residues tested for an arsenic-specific effect are indicated with arrows - W84C (pink), S112C (blue), and R113C (black). The lysine that is post-translationally modified with a lipoid acid is highlighted in red. (B) The percent increase of edited human cells that contain the W84C, S112C, or R113C amino acid change in DBT1 in the presence 5 µM arsenic trioxide relative to control conditions are shown. The number of reads in 5 µM arsenic trioxide for all replicates are significantly different from control conditions (Fisher’s exact test, p-value<0.011).
-
Figure 5—source data 1
Human cell line read data for CRISPR replacement experiment in 293 T cells.
(Used to generate Figure 5B).
- https://doi.org/10.7554/eLife.40260.069
-
Figure 5—source data 2
Results from Fisher’s exact test of human cell line read data for CRISPR replacement experiment in 293 T cells.
- https://doi.org/10.7554/eLife.40260.070
-
Figure 5—source data 3
Metabolite measurements from human cell line experiments.
- https://doi.org/10.7554/eLife.40260.071
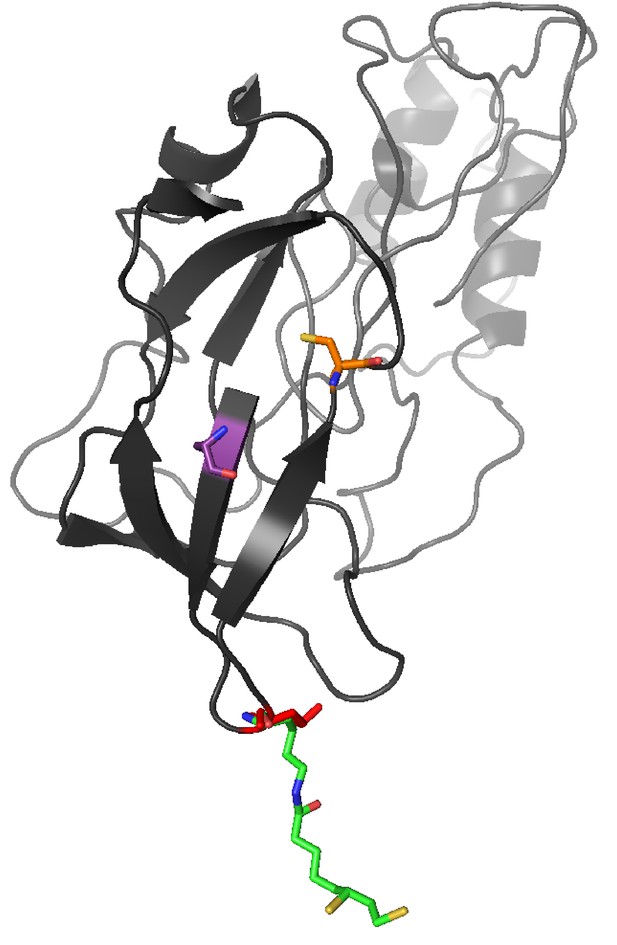
Three-dimensional homology model of C. elegans DBT-1 A three-dimensional homology model of C. elegans DBT-1 (black) aligned to human pyruvate dehydrogenase lipoyl domain (PDB:1Y8N) is shown.
The C78 residue that conferred resistance to arsenic trioxide is highlighted in orange, and the C65 residue is highlighted in purple. The C. elegans K71 residue is highlighted in red, and the human lipoylated lysine is highlighted in green.
-
Figure 5—figure supplement 1—source data 1
Tajima’s D of GWA mapping confidence interval.
(Used to generate Figure 5—figure supplement 1).
- https://doi.org/10.7554/eLife.40260.068
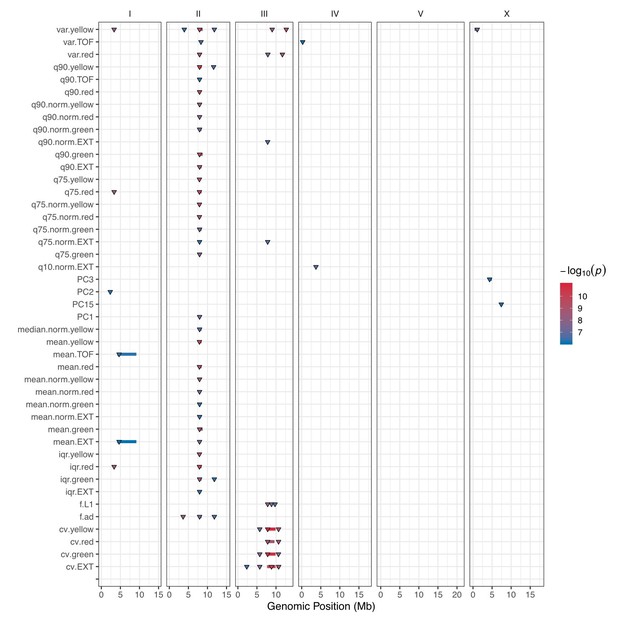
GWA mapping QTL summary.
All QTL identified by GWA mapping are shown. Traits are labeled on the y-axis, and the genomic position in Mb is plotted on the x-axis. Triangles represent the peak QTL position, and bars represent the associated QTL region of interest. Triangles and bars are colored based on the significance value, where red colors correspond to higher significance values.
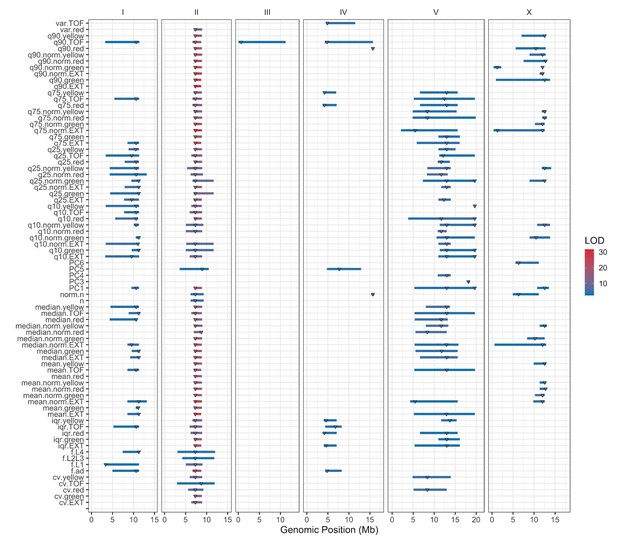
Linkage mapping QTL summary.
All QTL identified by linkage mapping are shown. Traits are labeled on the y-axis, and the genomic position in Mb is plotted on the x-axis. Triangles represent the peak QTL position, and bars represent the associated 1.5-LOD drop QTL confidence interval. Triangles and bars are colored based on the LOD score, where red colors correspond to higher LOD values.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | dbt-1 | NA | Wormbase:WBGene00014054 | |
| Strain, strain background (C. elegans) | N2 DBT-1(S78) | This paper | Andersen_Lab:ECA581 | dbt-1(ean15[C78S]) |
| Strain, strain background (C. elegans) | CB4856 DBT-1(C78) | This paper | Andersen_Lab:ECA590 | dbt-1(ean34[S78C]) |
| Strain, strain background (C. elegans) | Left NIL; CB4856 > N2 (II:5.75–8.02 Mb) | This paper | Andersen_Lab:ECA414 | eanIR188[II:5.75–8.02 Mb, CB4856 > N2] |
| Strain, strain background (C. elegans) | Right NIL; CB4856 > N2 (II:7.83–9.66 Mb) | This paper | Andersen_Lab:ECA434 | eanIR208[II:7.83–9.66 Mb, CB4856 > N2] |
| Sequence-based reagent | NIL Fd primer | This paper | Andersen_Lab:oECA609 | tttcacacaaaccatgcgct |
| Sequence-based reagent | NIL Rv primer | This paper | Andersen_Lab:oECA610 | actcgtctgctgggtattct |
| Sequence-based reagent | NIL Fd primer | This paper | Andersen_ Lab:oECA611 | tgtcttcgcacctttactcg |
| Sequence-based reagent | NIL Rv primer | This paper | Andersen_ Lab:oECA612 | cattcaagtcagggcgtatcc |
| Sequence-based reagent | Genotype C78S Edit | This paper | Andersen_Lab:oECA1163 | GAAGGAATTGCCGAAGTTCAGGTTAAG |
| Sequence-based reagent | Genotype C78S Edit | This paper | Andersen_Lab:oECA1165 | CCGTCATCTCCACAAAAAGCTTTATCTCTC |
| Sequence-based reagent | dbt-1 gRNA | This paper | Andersen_ Lab:crECA97 | CCATCTCCTGTAGATACGAC |
| Sequence-based reagent | N2 dbt-1 repair oligo | This paper | Andersen_ Lab:oECA1542 | CTTCCAGGTACGTGAAAGAAGGAGATACGATTTCGCAGTTCGATAAAGTCTGTGAAGTGCAAAGTGATAAAGCAGCAGTAACCATCTCCAGTAGATACGACGGAATTGTCAAAAAATTGTAAGTTTCTTCCTAA |
| Sequence-based reagent | CB4856 dbt-1 repair oligo | This paper | Andersen_ Lab:oECA1543 | TTAGGAAGAAACTTACAATT TTTTGACAATTCCGTCGTATCTACAGGAGATGGTTACTGCT GCTTTATCGCTTTGCACTTCACAGACTTTATCGAACTGCGAAATCGTATCTCCTTCTTTCACGTACCTGGAAG |
| Sequence-based reagent | dpy-10 repair oligo | Kim et al., 2014 | Andersen_Lab:crECA37 | CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT |
| Sequence-based reagent | dpy-10 gRNA | Kim et al., 2014 | Andersen_Lab:crECA36 | GCTACCATAGGCACCACGAG |
| Sequence-based reagent | Human gRNA S112C and R113C | This paper | Guide_1 used in RDA_74 | TCCATCATAACGACTAGTGA |
| Sequence-based reagent | S112C repair template | This paper | 1192 DBT1-repair-S112C | ATAGCATCTGTGAAGTTCAAAGTGATAAAGCTTCTGTTACAATCACTTGTCGTTATGATGGAGTCATTAAAAAACTCTATT |
| Sequence-based reagent | R113C repair template | This paper | 1193 DBT1-repair-R113C | ATAGCATCTGTGAAGTTCAAAGTGATAAAGCTTCTGTTACAATCACTAGTTGTTATGATGGAGTCATTAAAAAACTCTATT |
| Sequence-based reagent | Fwd PCR C | This paper | 1188 DBT1-PCR-C | TtgtggaaaggacgaaacaccgAGAAGGAGATACAGTGTCTCAGT |
| Sequence-based reagent | Fwd PCR D | This paper | 1189 DBT1-PCR-D | TtgtggaaaggacgaaacaccgTGTCTCAGTTTGATAGCATCTGTG |
| Sequence-based reagent | Human gRNA W84C | This paper | Guide_2 used in RDA_75 | TCTTTTAGGTATGTAAAAGA |
| Sequence-based reagent | W84C repair template | This paper | 1195 DBT1-repair-W84C-v2 | GACTGTTTCCATAAAAGTGTCTCATTTCTTTTTCTTTTAGTTATGTGAAGGAAGGAGATACAGTGTCTCAGTTTGATAGCAT |
| Sequence-based reagent | Fwd PCR A | This paper | 1186 DBT1-PCR-A | TtgtggaaaggacgaaacaccgGCATGGCATTTACATCCTTAATATGAT |
| Sequence-based reagent | Fwd PCR B | This paper | 1187 DBT1-PCR-B | TtgtggaaaggacgaaacaccgCCTTAATATGATCTGTACTTATGACTGTTT |
| Sequence-based reagent | Rev PCR 1 | This paper | 1190 DBT1-PCR-Rev1 | TctactattctttcccctgcactgtCTACTAATGGCTTCCCCACAT |
| Sequence-based reagent | Rev PCR 2 | This paper | 1191 DBT1-PCR-Rev2 | TctactattctttcccctgcactgtCAATACCTTTTAAAGCTTCCGTTTCTAT |
| Transfected construct (Homo Sapiens) | S112C and R113C Cas9-sgRNA plasmid | This paper | p1054 | |
| Transfected construct (Homo Sapiens) | W84C Cas9-sgRNA plasmid | This paper | p1052 |
Additional files
-
Supplementary file 1
Plasmid used for editing human cells with the S112C and R113C edits.
- https://doi.org/10.7554/eLife.40260.072
-
Supplementary file 2
Plasmid used for editing human cells with the W84C edit.
- https://doi.org/10.7554/eLife.40260.073
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40260.074





