H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes
Figures
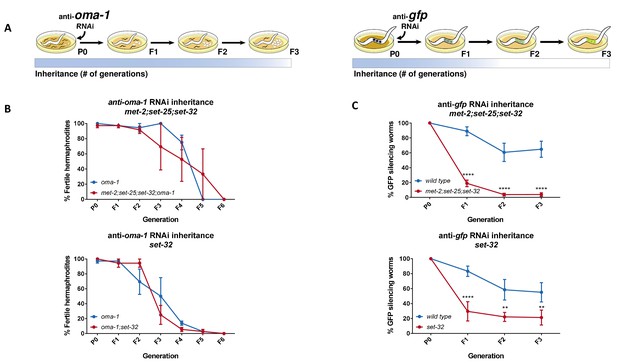
Heritable RNAi responses against the oma-1 and gfp genes have different requirements for H3K9me3 methyltransferases.
(A) Scheme depicting the different requirements for H3K9 methyltransferases in RNAi inheritance responses aimed at different genes. In the parental generation, worms are exposed to RNAi by growing on plates seeded with dsRNA-producing bacteria. Next, worms are transferred to plates seeded with control bacteria (that do not express dsRNA) to lay the eggs the next generation. (left) Only worms that inherit small RNAs that silence the temperature-sensitive dominant allele of oma-1 can hatch. Heritable RNAi responses aimed against the endogenous oma-1 gene do not require H3K9me3 methyltransferases. (right) Inheritance of anti-gfp small RNAs lead to heritable silencing of the gfp transgene (Pmex-5::gfp::h2b transgene). Heritable RNAi responses aimed against the foreign gfp gene strongly depends on H3K9me3 methyltransferases. (B) Inheritance of anti-oma-1 RNAi response in H3K9me3 methyltransferase mutants. The percentage of fertile worms per replicate and generation is presented (N = 12, three biological replicates). (upper panel) RNAi inheritance dynamics in met-2;set-25;set-32;oma-1 mutants compared to oma-1 mutants. (lower panel) RNAi inheritance dynamics in set-32;oma-1 mutants compared to oma-1 mutants. (C) Inheritance of anti-gfp RNAi response in H3K9me3 methyltransferase mutants. In each generation the percentage of worms silencing a germline expressed GFP transgene is presented (N > 60, five replicates). (upper panel) RNAi inheritance dynamics in met-2;set-25;set-32 triple mutants. (lower panel) RNAi inheritance dynamics in set-32 single mutants. Error bars represent standard error of mean. *p-value<0.05, **p-value<0.005, ***p-value<0.001, ****p-value<0.0001, Two-way ANOVA, Sidak's multiple comparisons test.

Weak anti-gfp heritable RNAi silencing of gfp in methyltransferases mutants.
Scatter column representation of the normalized GFP germline expression levels of RNAi treated animals across generations. The GFP expression levels were normalized to the expression levels of the corresponding (same generation and repeat) untreated control animals (three independent biological repeats). Each dot represent the normalized germline GFP expression level of a specific animal (dots representing different animals with similar expression levels may be visually indistinguishable). (A) Heritable anti-gfp silencing dynamics in met-2;set-25;set-32 triple mutants (Red) and wild type animals (Blue, in total, the germlines of 1350 worms were quantified). (B) Heritable anti-gfp silencing dynamics in set-32 mutants (Red) and wild type animals (Blue, in total, the germlines of 1480 worms were quantified). Error bars represent standard deviations.

SET-32 is required for the strong heritable RNAi-induced silencing of gfp in met-2 mutants.
(A) Inheritance of anti-gfp RNAi response in set-32;met-2 double mutants and wild type is shown. In each generation, the percentage of worms silencing a germline expressed GFP transgene is presented (N > 60, three biological replicates). *p-value<0.05, **p-value<0.005, ***p-value<0.001, Two-way ANOVA, Sidak's multiple comparisons test. Error bars represent standard error of mean. (B) Table summarizing the genetic interaction between the H3K9 methyltransferases MET-2, SET-25 and SET-32 and their requirement for RNAi-induced H3K9me3 and heritable silencing of gfp.
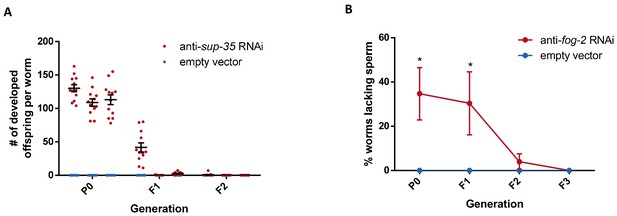
RNAi-induced silencing of fog-2 or sup-35 is not inherited transgenerationally.
(A) Inheritance of RNAi-induced silencing of sup-35. Silencing of the sup-35 gene by anti-sup-35 RNAi allows the temperature-sensitive pha-1 mutants to develop in restrictive temperatures. The number of developed progeny per worm is indicated (three biological repeats, N = 12). Error bars represent standard error of mean. (B) Inheritance of RNAi-induced silencing of fog-2. Silencing of the fog-2 gene depletes the worm of sperm, and causes stacking of unfertilized oocytes. The average percentage of worms lacking sperm is shown (three biological repeat, N > 30 per repeat). *p-value<0.05, Two-way ANOVA, Sidak's multiple comparisons test.
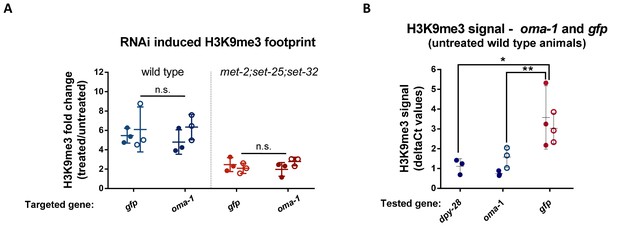
The fold change in RNAi-induced H3K9me3 on oma-1 and gfp is comparable.
(A) The RNAi-induced H3K9me3 footprint on the RNAi-targeted genes. The fold change in H3K9me3 levels in F1 progeny of animals exposed to RNAi versus untreated control animals. The H3K9me3 footprint levels were assessed using a qPCR quantification of ChIP experiments conducted with both wild type (left) and met-2;set-25;set-32 mutants (right). Filled or empty circles represent qPCR data obtained using two different primer sets that span different parts of the examined locus. (B) H3K9me3 levels on the gfp and oma-1 genes in naive untreated wild type animals. The deltaCt numbers used to obtain the fold change values were calculated using the eft-3 gene as an endogenous control. The presented data were obtained from three biological replicates. The levels of gfp and dpy-28 H3K9me3 signal in wild type animals are adapted from raw data from our previous publication (Lev et al., 2017). Two-way ANOVA, Sidak's multiple comparisons test. **p-value<0.005. Error bars represent standard deviations.
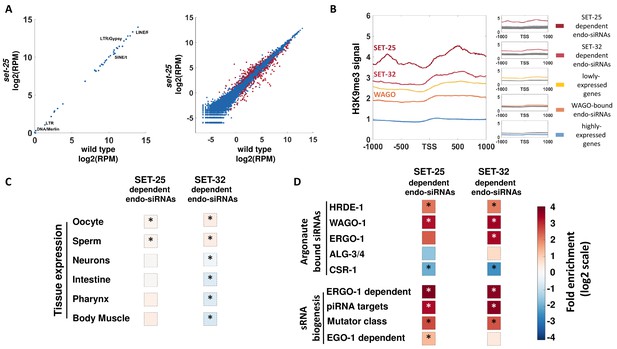
A genome-wide analyses of endo-siRNAs that depend on SET-25 or SET-32.
(A) An expression analysis of endo-siRNAs targeting transposons and repetitive elements classes (left panel) or protein-coding genes (right panel). Shown are the expression values as log2 of number of Reads per Million (RPM) in set-25 mutants (y-axis) compared to wild type animals (x-axis). Gene targets of endo-siRNAs, which display significant differential expression (analyzed with Deseq2, adjusted p-value<0.1) are marked in Red (B) An analysis of H3K9me3 signals (based on published data from McMurchy et al., 2017) on differet sets of gens: highly expressed genes (top 10%, Blue), lowly expressed genes (top 10%, Yellow) and gene targets of endo-siRNAs that depend on SET-25 (based on Lev et al., 2017, Red), SET-32 (based on Kalinava et al., 2018), Dark Red) or endo-siRNAs associated with WAGOs (HRDE-1,WAGO-1, and ERGO-1, Light Red). H3K9me3 signal is aligned according to gene's Transcription Start Sites (TSS), and the regions of 1000 base pairs upstream and downstream of the TSS are shown on the x axis. The y axis shows the averaged signal of the H3K9me3 modification as a function of distance from the TSS. For statistical analysis, control data sets (shown in Gray) were created by sampling the H3K9me3 levels of randomly selected gene sets of the same size as the examined gene list. (C and D) An enrichment analysis of genes with significantly lowered levels of endo-siRNAs targeting them in set-25 and set-32 mutants compared to wild type. Fold enrichment values (log2 scale) are color coded. (C) An enrichment analysis for expression in specific tissues. (D) An enrichment analysis for different small RNA pathways. The p-values were calculated using 10,000 random gene sets identical in their size to the examined endo-siRNA-target gene list. Asterisk denotes statistically significant enrichment values (p-value<0.05).
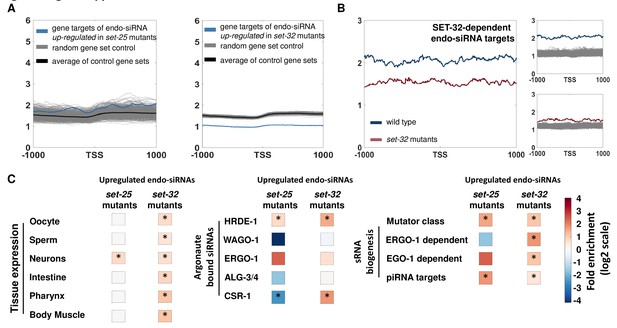
An analysis of genes targeted by endo-siRNAs upregulated in set-25 and set-32 mutants.
(A) An analysis of H3K9me3 signals (based on published data from McMurchy et al., 2017) on gene targets of endo-siRNAs up-regulated in set-25 mutants (based on Lev et al., 2017, left panel) and in set-32 mutants (based on Kalinava et al., 2018). All genes are aligned according to their Transcription Start Sites (TSS), and the regions of 1000 base pairs upstream and downstream of the TSS are shown on the x axis. The y axis shows the averaged signal of the H3K9me3 modification as a function of distance from the TSS. (B) An analysis of H3K9me3 signals (based on Kalinava et al., 2018) on gene targets of SET-32 dependent endo-siRNAs. Genes are aligned according to their TSS, and the regions of 1000 base pairs upstream and downstream of the TSS are shown on the x axis. The y axis shows the averaged signal of the H3K9me3 modification in wild type (Blue) and set-32 mutants (Red). For statistical analysis control data sets (Gray) were created by sampling the H3K9me3 levels of randomly selected gene sets of the same size as the examined gene list. (C) An enrichment analysis for expression in specific tissues or association with different small RNA pathways. Fold enrichment values are color coded. For statistical analysis, p-values were calculated using 10,000 random gene sets identical in their size to the examined gene set. Asterisk denotes statistically significant enrichment values (p-value<0.05).
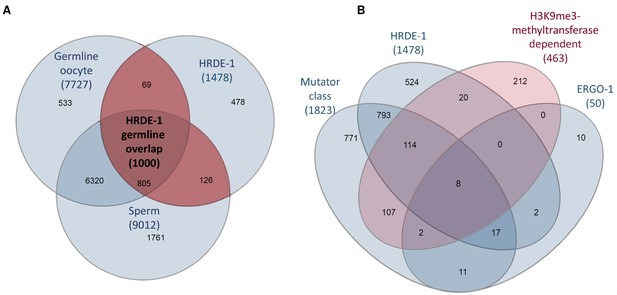
An analysis of overlap between different endo-siRNA target gene sets.
(A) A Venn diagram analysis of the overlap between gene targets of HRDE-1-bound endo-siRNAs (Buckley et al., 2012) and germline expressed genes (Ortiz et al., 2014). (B) A Venn diagram comparison of gene targets of endo-siRNAs that depend on either SET-25 or SET-32 and genes targeted by known endo-siRNA pathways: HRDE-1 (Buckley et al., 2012), Mutator pathway (Phillips et al., 2014), ERGO-1 dependent small RNA targets (Vasale et al., 2010) and endo-siRNAs dependent on either SET-25 (Lev et al., 2017) or SET-32 (Kalinava et al., 2018).
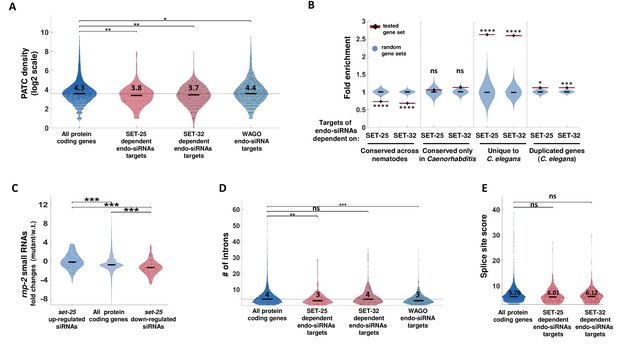
SET-25-dependent endo-siRNAs target newly evolved genes.
(A) A PATC density analysis for SET-25 and SET-32-dependent endo-siRNAs gene targets. The PATC density values (obtained from Frøkjær-Jensen et al., 2016) are presented for all protein-coding genes, gene targets of endo-siRNAs that depend on SET-25 or SET-32 and gene targets of endo-siRNAs associated with WAGO small RNA pathways (HRDE-1,WAGO-1 or ERGO-1). **p-value<0.005, *p-value<0.05, Wilcoxon rank sum test. For clarity of display, values are shown in log2 scale (after addition of 1). The median (black line) and average levels (numbers) of PATC density levels of each plot are indicated (log2 scale). (B) An enrichment analysis of genes conserved at different levels and duplicated genes amongst gene targets of SET-25- and SET-32-dependent endo-siRNAs. The gene sets were generated based on the homology field in WormBase that details the orthologs and paralogs of each nematode gene. We defined a duplicated gene as a gene that has a paralog in C. elegans. We define genes unique to C. elegans as genes that lack an ortholog amongst the nematode species we examined (see Materials and methods). For statistical analysis, control enrichment values were obtained from 10,000 random gene sets with the same size as the examined endo-siRNA-target gene list. ****p-value<0.0001,***p-value<0.001, *p-value<0.05 (C) An analysis of endo-siRNAs fold changes in rnp-2 mutants for genes targets of endo-siRNAs downregulated or upregulated in set-25 mutants or all genes. All p-values<0.001, Wilcoxon rank sum test. (D) An analysis of intron numbers of gene targets of SET-25- and SET-32-dependent endo-siRNAs and WAGO-associated endo-siRNAs compared to all protein-coding genes. In cases of genes that have more than one transcript, the average intron value is used. The median intron number of each plot is indicated (log2 scale). ***p-value<0.001,**p-value<0.005, Wilcoxon rank sum test. (E) An analysis of splicing motif divergence score (based on Newman et al., 2018) of gene targets of SET-25 and SET-32-dependent endo-siRNAs, WAGO associated endo-siRNAs and all protein-coding genes. The median score levels of each plot are indicated. p-value>0.05, Wilcoxon rank sum test.
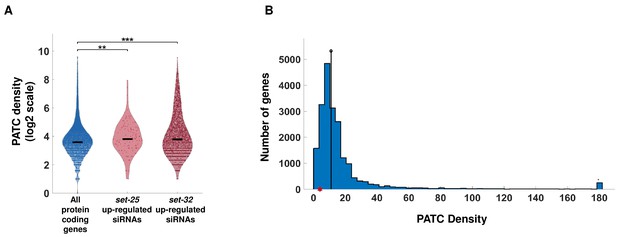
An analysis PATC density of the oma-1 gene and of targets of endo-siRNAs upregulated in set-25 and set-32 mutants.
(A) A PATC density analysis for genes targeted by endo-siRNAs upregulated in set-25 and set-32 mutants. The PATC density values (obtained from Frøkjær-Jensen et al., 2016) are presented for all protein-coding genes, gene targets of endo-siRNAs that are upregulated in set-25 and set-32 mutants (log2 scale). ***p-value<0.001, **p-value<0.005, Wilcoxon rank sum test. (B) A histogram of PATC density of all of C. elegans genes (Blue) and the PATC density of the oma-1 gene (Red dot). The median PATC density of all genes is 11.00 (marked with black vertical line), 243 points counted in the rightmost bin have values greater than 178.47.

Gene targets of endo-siRNAs that depend on SET-25 or SET-32 are preferably targeted by small RNAs on intron-exon junctions compared to all protein coding genes.
(A) An analysis of endo-siRNAs fold changes in rnp-2 mutants for genes targets of endo-siRNAs downregulated or upregulated in set-32 mutants or all genes. ***p-values<0.001, Wilcoxon rank sum test. (B) The lower intron numbers found in genes targeted by SET-25 dependent endo-siRNAs are not explained by smaller coding sequence sizes. An analysis of coding sequence length of genes targeted by SET-25 dependent endo-siRNAs and all protein-coding genes. p-value=0.8673, Wilcoxon rank sum test. This figure relates to Figure 4D. (C and D) The proportion of genes with different ratios of small RNAs aligned to Intron-Exon-junction/Exon Exon-junction (based on Newman et al., 2018) are presented. For statistical analysis, control proportion values (Gray) were obtained from 10,000 random gene sets (each set was identical in size to the examined gene set), ****p-value<0.0001,*p-value<0.05. The proportion values of the examined gene set are shown (Red) for gene targets of SET-25 dependent endo-siRNAs (C) and SET-32 dependent endo-siRNAs (D). (E) An analysis of small RNAs aligned to the studied gfp construct. Shown are small RNAs aligned to the gfp construct used in this study in naive wild type animals (Lev et al., 2017). A scheme of the gfp transgene construct used in this study is also presented. The scheme denotes all the relevant endogenous and foreign elements included in the construct. Information regarding the PATC on the Pmex-5 promoter was obtained from Frøkjær-Jensen et al. (2016).
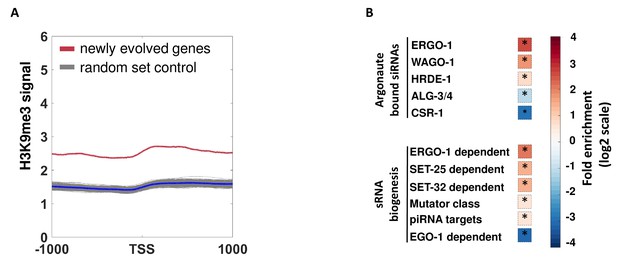
An analysis of endo-siRNAs targeting and H3K9me3 levels of newly evolved genes.
(A) An analysis of H3K9me3 levels (based on published data from McMurchy et al., 2017) of newly acquired genes in C. elegans. The newly acquired gene set was generated based on the ‘Homology’ field of the C. elegans genes in WormBase. As a control, shown are the H3K9me3 signature of 500 random gene sets identical in size to the examined gene set. (B) An enrichment analysis for newly acquired genes, having no orthologues inother nematodes amongst different endo-siRNAs pathways. Fold enrichment values are color coded. Asterisk denotes statistically significant enrichment values (p-value<0.005). P-values were calculated using 10,000 random gene sets identical in their size to the examined gene set.
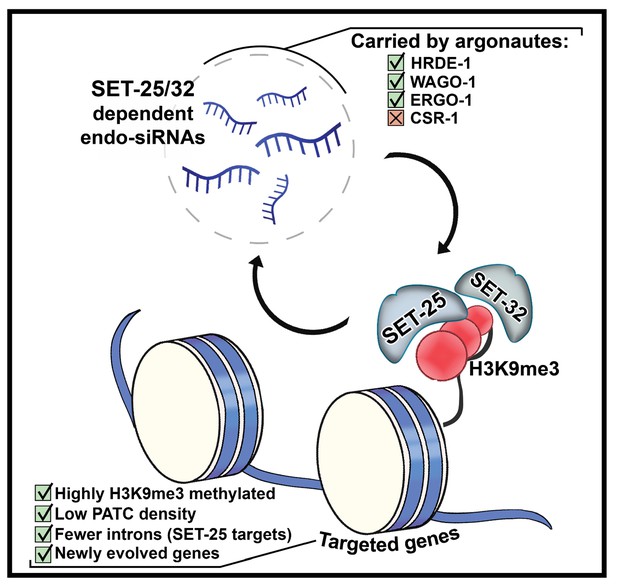
Scheme characterizing H3K9me3 methyltransferase-dependent endo-siRNAs and their targets.
SET-25-dependent and SET-32-dependent endo-siRNAs are enriched with small RNAs known to be carried by the argonautes HRDE-1, WAGO-1 and ERGO-1 but not CSR-1. SET-25-dependent and SET-32-dependent endo-siRNAs targets are enriched with newly evolved genes, have fewer PATC sequences, and are marked with higher levels of H3K9me3. Targets of SET-25-dependent but not SET-32-dependent endo-siRNAs bear fewer introns.
Tables
| Reagent type | Designation | Source of reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (E. coli) | OP50 | (Brenner, 1974) | Op50 RRID:WB-STRAIN:OP50 | |
| Strain (E. coli) | anti-gfp RNAI bacteria | This study | ||
| Strain (E. coli) | anti-oma-1 RNAi bacteria | Ahringer RNAi library (Kamath and Ahringer, 2003) | 4-4-3-C01 | |
| Strain (E. coli) | anti-sup-35 RNAi bacteria | Vidal RNAi library (Rual et al., 2004) | 11006-E2 | |
| Strain (E. coli) | anti-fog-2 RNAi bacteria | Vidal RNAi library (Rual et al., 2004) | 10011-C3 | |
| Strain (C. elegans) | N2 | CGC | N2 RRID:WB-STRAIN:N2_(ancestral) | |
| Strain (C. elegans) | set-32(ok1457) ; oma-1(zu405) | CGC | BFF25 | |
| Strain (C. elegans) | mjIs134[pmex-5::gfp::h2b::tbb-2] | Erik Miska's lab (Univeristy of Cambridge) | SX1263 | |
| Strain (C. elegans) | oma-1(zu405) | CGC | TX20 | |
| Strain (C. elegans) | pha-1(e2123) | CGC | GE24 | |
| Strain (C. elegans) | set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF24 | |
| Strain (C. elegans) | met-2(n4256);set-25(n5021);set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF26 | |
| Strain (C. elegans) | met-2(n4256);set-25(n5021);set-32(ok1457);oma-1(zu405) | This study | BFF27 | |
| Strain (C. elegans) | met-2(n4256);set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF28 | |
| Antibody | H3K9me3 | Abcam | RRID:AB_306848 | |
| Software, algorithm | GraphPad Prism | https://www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 | |
| Software, Algorithm | Image J | Opensource: https://imagej.nih.gov/ij/ | RRID:SCR_003070 | |
| Software, Algorithm | MATLAB MathWorks | https://www.mathworks.com/ | RRID:SCR_016651 |
Additional files
-
Supplementary file 1
Sequence of anti-gfp RNAi targeting dsRNA used in this study.
- https://doi.org/10.7554/eLife.40448.015
-
Supplementary file 2
List of gene targets of endo-siRNAs that depend on SET-25 or SET-32 and their conservation.
The table includes all the C. elegans genes we examined (20,447 genes). Each gene is marked for being: A. a target of SET-25 dependent endo-siRNAs, B. a target of SET-32 dependent endo-siRNAs. C. Conserved among nematodes, D. Conserved only in Caenorhabditis, E. Unique to C. elegans.
- https://doi.org/10.7554/eLife.40448.016
-
Supplementary file 3
Fold change enrichment values for gene targets of endo-siRNAs that depend on SET-25 or SET-32.
The table lists the fold change enrichment values and the matching p-values found in gene targets of SET-25 and SET-32 dependent endo-siRNAs for other genes sets discussed in this manuscript.
- https://doi.org/10.7554/eLife.40448.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40448.018





