RalB directly triggers invasion downstream Ras by mobilizing the Wave complex
Figures
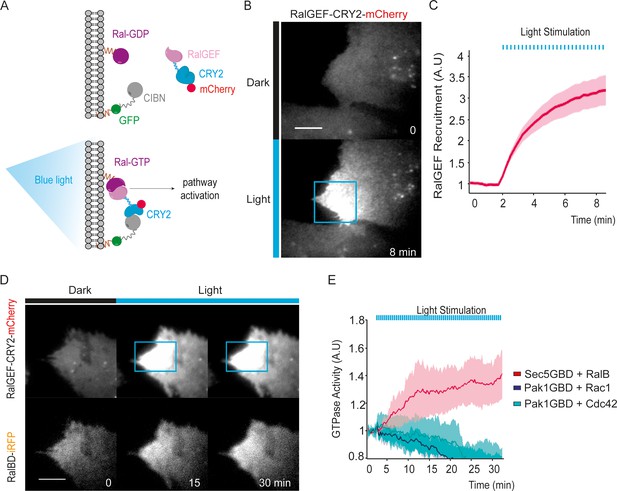
Optogenetic control of Ral activation.
(A) The OptoRal strategy. Upon blue light stimulation the RalGEF domain of RGL2, fused to CRY2, is recruited to the plasma membrane following the interaction between CRY2 and CIBN, which is targeted to the plasma membrane by a CAAX prenylation motif. mCherry and GFP fluorescent proteins were used to monitor expression and localization of RalGEF-CRY2 and CIBN, respectively. After recruitment, the RalGEF induces activation of endogenous Ral. (B) Representative RalGEF-CRY2-mCherry recruitment. The fluorescent RalGEF-CRY2-mCherry fusion protein was imaged by TIRF microscopy before illumination (dark) and 8 min after blue light stimulation inside the blue square area (100 ms pulses every 15 s). Scale bar, 10 μm. See Video 1 for the entire sequence. (C) Quantification of RalGEF-CRY2-mCherry recruitment. Average time course of the fold increase of mCherry fluorescence, that is RalGEF recruitment, inside the illuminated square area, is calculated from n = 20 cells from three independent experiments. Lines represent the mean, shaded regions represent the standard deviation (SD). (D) Representative Ral activation. The fluorescent RalGEF-CRY2-mCherry and Sec5GBD-iRFP (reporter of Ral activity) fusion proteins were simultaneously imaged by TIRF microscopy. Scale bar, 10 μm. See Video 2 for the entire sequence. (E) Light activates RalB, but not Rac1 or Cdc42. OptoRal cells were transiently transfected to express: Sec5GBD-iRFP (reporter of Ral activity) with GFP-RalB (red line), Pak1GBD-iRFP (reporter of Rac1/Cdc42 activity) with GFP-Rac1 (blue line) or Pak1GBD-iRFP with GFP-Cdc42, respectively (light blue line). Average time course of the fold increase of iRFP fluorescence, that is Ral or Rac1/Cdc42 activities, inside the illuminated square area, is calculated from n = 6 cells per condition from three independent experiments. Lines represent the mean, shaded regions represent the standard deviation (SD).
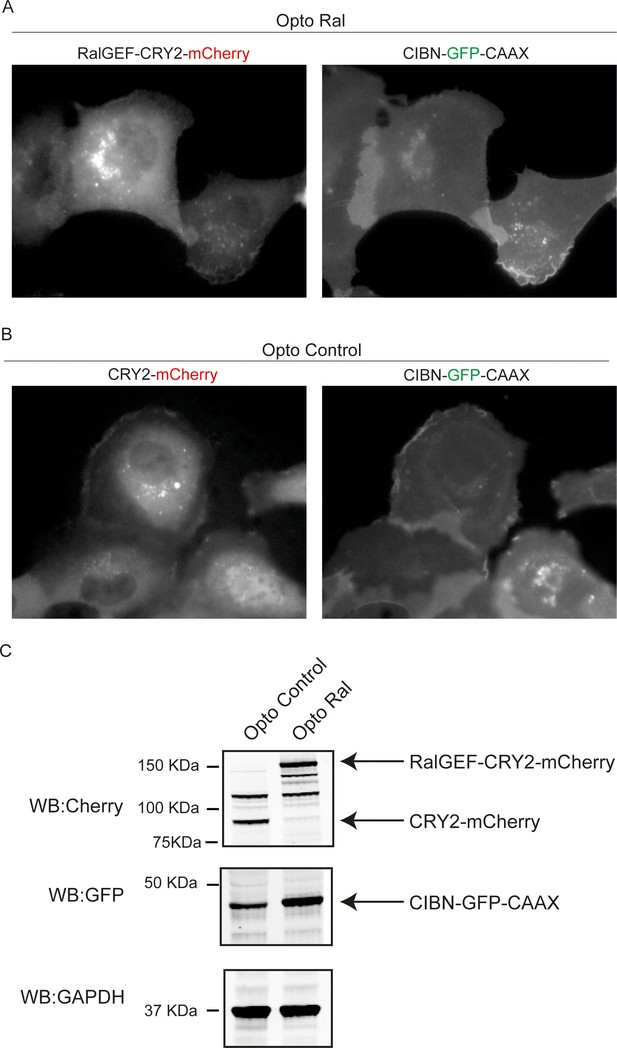
Optogenetic pair expression.
(A) Representative images of RalGEF-CRY-mCherry and CIBN-GFP-CAAX signals by epifluorescence microscopy before blue light stimulation in OptoRal cells. (B) Representative images of CRY-mCherry and CIBN-GFP-CAAX signals by epifluorescence microscopy before blue light stimulation in OptoControl cells. (C) Representative western blots for Cherry and GFP from cell lysates of OptoControl and OptoRal cells. GAPDH is used as loading control. The 110 kDa band in the Cherry western blot is non-specific.
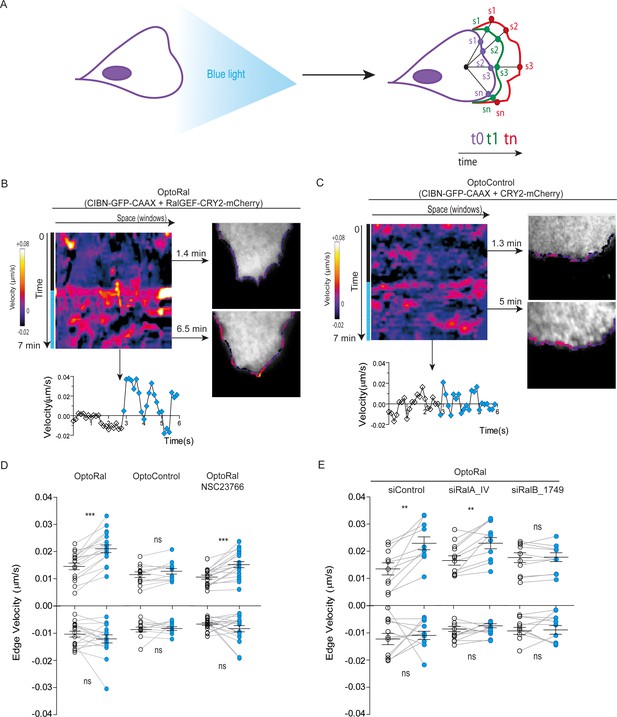
Local Ral activation induces protrusiveness.
(A) Cell edge dynamics inside the illuminated area were measured using an automated method. (B) Edge dynamics velocity map of a representative OptoRal cell before and after illumination (dotted blue line). See Video 3 for the entire sequence. The color-coded map shows the velocity measurements for each sampled edge window (space, horizontal axis) and for each time point (time, vertical axis). Positive velocities (i.e. protrusion) are represented as warm color, negative velocities (i.e. retraction) as cold colors. As example of velocity profile over time, the velocity measurements are shown for a selected point in space (window 71) in the lower panel; the stimulation of edge velocity is visible immediately after light illumination. As examples of velocity profiles over the space dimension, the velocity measurements are shown in the right panels using the color-code along the cell edge for two selected time points, 1.4 min and 6.5 min, dark and light conditions, respectively. (C) Edge dynamics velocity map of a representative OptoControl cell before and after illumination. See Video 4 for the entire sequence. The velocity profiles over time (window 77, lower panel) or over space (1.3 min and 5 min, right panels) show that illumination did not induce any change of edge velocity. (D) Quantification of edge velocities before and after illumination. Positive velocities (i.e. protrusion) and negative velocities (i.e. retraction) are analysed separately. Each white dot represents the mean of all velocity measurements (all time and space points) before illumination. Each blue dot represents the mean of all velocity measurements (all time and space points) after illumination. Measurements from a same cell are connected by lines. Illumination stimulates positive edge velocities in OptoRal cells (left) but not in OptoControl cells (centre). Inhibition of Rac with NSC23766 (100 µM, treatment started 1 hr before experiment) did not impair stimulation by light of positive edge velocities in OptoRal cells (right). Bars represent mean of n = 18 cells per condition ±SEM from three independent experiments. *** indicates p<0.001, ** indicates p<0.01 and ns indicates not-significant, using Wilcoxon signed-rank test for paired measurements (same cell pre- and post-illumination). There was no significant difference in the delta velocities pre- and post-illumination when comparing presence and absence of the NSC23766 inhibitor (using Student t-test, not shown). There was no significant difference in the pre-illumination edge velocities when comparing OptoRal and OptoControl cells (using Student t-test, not shown). (E) RalB depletion, but not RalA depletion, impairs stimulation of positive edge velocities in OptoRal cells. OptoRal cells were transfected with the indicated siRNAs and analysed as in panel D. n = 10–11 cells per condition.
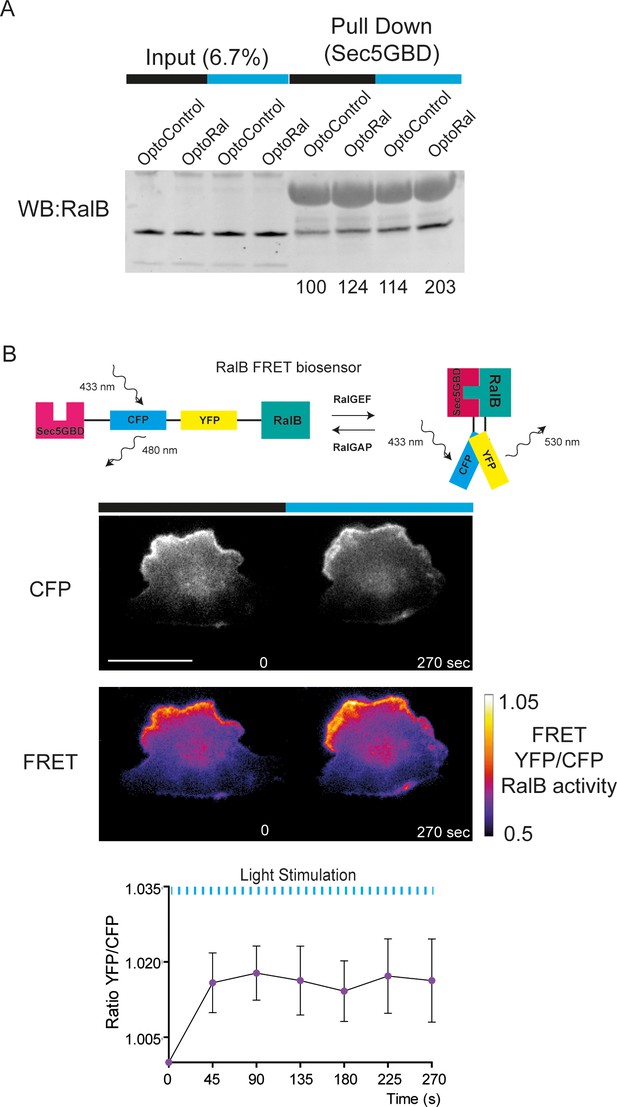
Additional evidences that light stimulates RalB activity in OptoRal cells.
(A) Pull-down assay of RalB-GTP in dark and after blue light stimulation (5 min), in OptoControl and OptoRal cells, using Sec5GBD beads. The quantifications express the RalB-GTP/total RalB ratios, normalized for OptoControl dark condition. Cells were starved overnight in DMEM supplemented 0.2% FBS before the assay. For dark conditions, cells were kept in a dark box to avoid daylight exposure and lysed in a dark room. For light conditions, cells were exposed to 488 nm excitation illumination under an epifluorescence microscope for 5 min prior lysis. (B) Visualization of RalB activity by FRET. OptoRal cells, expressing a RalB FRET-based biosensor, were imaged by FRET miscoscopy; in such setting, since the CFP stimulation results in optogenetics activation, the first time point is taken as dark condition. The graph shows the average ±SD whole cell FRET measurements from n = 3 cells. Scale bar, 10 µm.
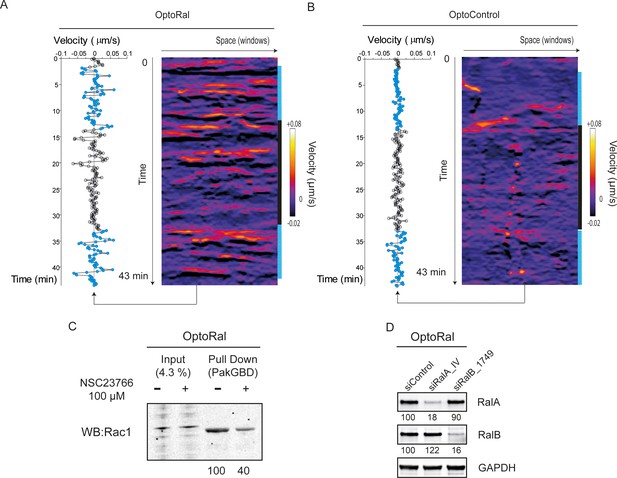
Reversibility and repeatability of edge velocity stimulation by light. Validation of Rac1 inhibition and of RalA and RalB depletion.
(A) Edge dynamics velocity maps of OptoRal cells upon alternation of dark and light periods. Pulsed light stimulation was alternated with periods of dark as following: 3 min dark, 10 min light, 20 min dark, and 10 min light. On the left, the velocity measurements over the time are shown for a selected space point (window 52). The stimulation of edge velocity repeatedly occurs immediately after light illumination. Upon switch off to dark, the reversion to base-line edge velocity occurs within 10 min. (B) Edge dynamics velocity maps of OptoControl cells upon alternation of dark and light periods. No matter light or dark, only small stochastic fluctuations of edge velocity are observed. (C) Validation of Rac1 inhibition by NSC23766 treatment. OptoRal cells were incubated 1 hr with 100 µM NSC23766 prior to cell lysis. Cell lysates were subjected to Rac1-GTP pull-down assay using Pak1GBD beads. The quantifications express the Rac1-GTP/total Rac1 ratios, normalized for control condition without treatment. (D) Validation of Ral depletions for cell edge dynamics in OptoRal cells. Representative western blots from cell lysates prepared 72 hr after transfection with the indicated siRNA. GAPDH was used as loading control. Quantifications of Ral depletion efficiencies, calculated as Ral/GAPDH and normalized for siControl, are shown below the WBs.
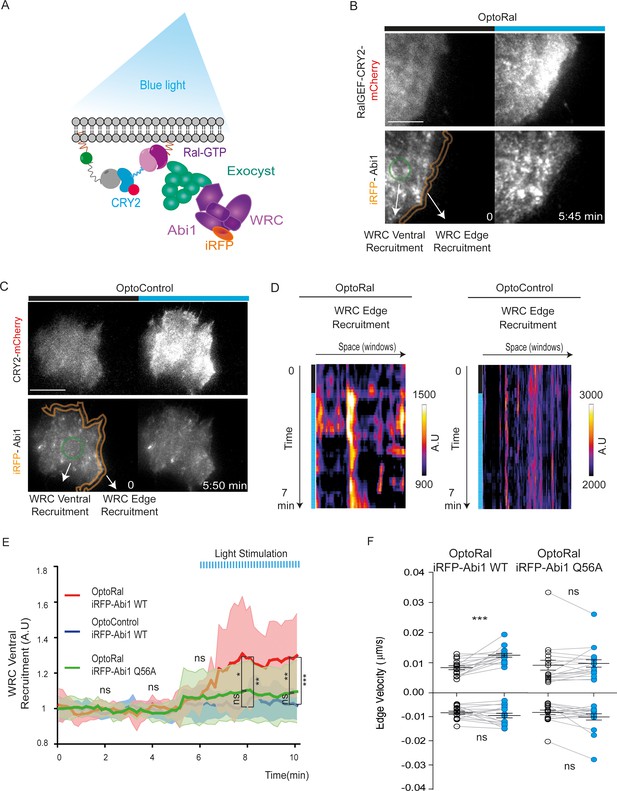
Local Ral activation induces WRC complex recruitment at the edge and ventral plasmamembrane via the exocyst.
(A) Working model. Upon Ral activation, exocyst is assembled and recruited at the plasma membrane, functioning as transporter for the WRC complex. The model was tested by monitoring the recruitment of WRC by TIRF microscopy using a fluorescent-tagged Abi1 subunit of WRC (iRFP-Abi1). (B) Representative iRFP-Abi1 recruitment (reporter of WRC localization) in OptoRal cells. The fluorescent RalGEF-CRY2-mCherry and iRFP-Abi1 fusion proteins were simultaneously imaged with TIRF microscopy before and 7 min after blue light stimulation. Scale bar, 10 μm. See Video 5 for the entire sequence. The variations of iRFP fluorescence were measured both at the ventral compartment (green circle) and at the leading edge (orange lines) of the cells. (C) Representative absence of light-dependent iRFP-Abi1 recruitment in OptoControl cells. Scale bar, 10 μm. See Video 6. There is an increase of mCherry but not of iRFP fluorescence after illumination. (D) Color-coded maps show the WRC edge recruitment in the OptoRal cell of panel B and in the OptoControl cell of panel C, calculated across a band with a width of 1.12 µm, before and after illumination, for each sampled edge window (space, horizontal axis) and for each time point (time, vertical axis). In the OptoRal cell (left), changes to warmer colors after illumination indicate increased WRC recruitment. In the OptoControl cell (right), there are no changes of WRC edge recruitment after illumination. (E) Quantification of ventral iRFP-Abi1 recruitment: comparison of OptoRal iRFP-Abi1 wild-type (WT), OptoControl iRFP-Abi1 wild-type (WT), and OptoRal iRFP-Abi1Q56A cells. Average time course of the fold increase of iRFP fluorescence, that is WRC recruitment, is calculated from n = 15 cells per condition from four independent experiments. Lines represent the mean, shaded regions represent the standard deviation (SD). Statistical comparison of the three curves was done using Student t-test at 2, 4, 6, 8, 10 min time points: *p<0.05, **p<0.01, ***, p<0.001, ns not-significant. Only the OptoRal iRFP-Abi1 wild-type curve diverges after light stimulation (at 6 min). (F) Quantification of edge velocities before and after illumination: comparison Abi1 wild-type (WT) versus Abi1Q56A in OptoRal cells. See legend Figure 2D for details. Illumination stimulates positive edge velocities in OptoRal cells expressing Abi1 WT (left) but not in OptoRal cells expressing Abi1Q56A (right). Bars represent mean of n = 15 cells per condition ±SEM from three independent experiments. *** indicates p<0.001 and ns indicates not-significant, using Wilcoxon signed-rank test for paired measurements (same cell pre- and post-illumination).
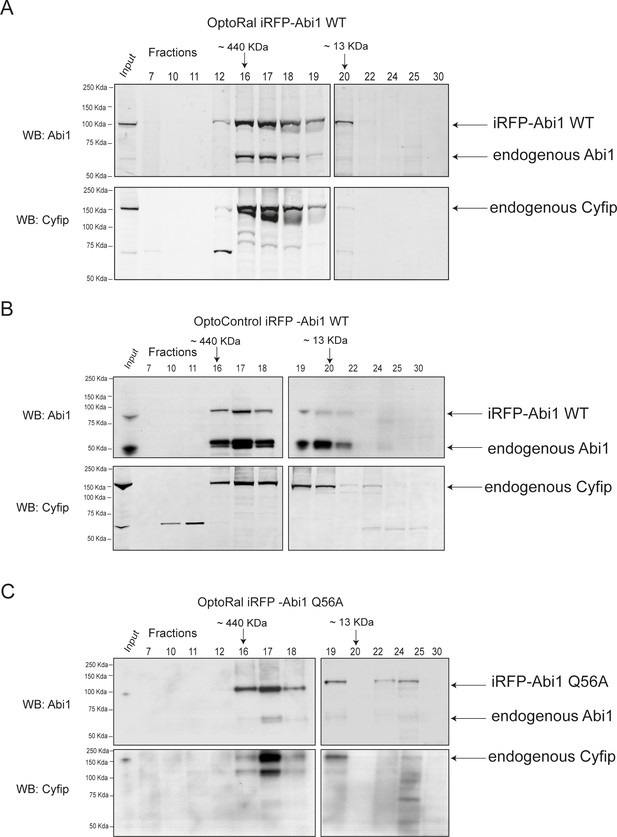
Size exclusion chromatography shows incorporation of exogenous iRFP-Abi1 (both wild-type and Q56A) into the whole WRC complex.
Total lysates from OptoRal cells expressing iRFP-Abi1 WT (A), from OptoControl cells expressing iRFP-Abi1 WT (B), or from OptoRal cells expressing iRFP-Abi1Q56A (C) were subjected to analytical size exclusion chromatography on a Superose 610/300 GL column. Input and fractions were analyzed by western blot using anti-Abi and anti-Cyfip antibodies. Exogenous iRFP-Abi fusion proteins co-eluted with endogenous Cyfip in the high molecular weight fractions, confirming that the fusion protein is incorporated in the WRC complex.
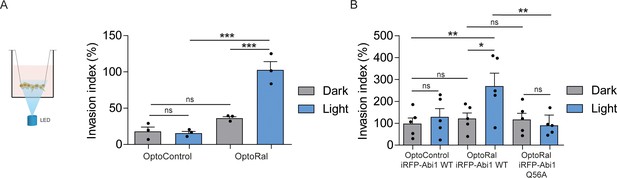
Activation of Ral triggers invasion of non-transformed cells.
(A) Transwell invasion assay with OptoControl and OptoRal cells, in the dark or under light stimulation. Graph shows the mean ±SEM from three independent experiments. In the graph the results of one-way ANOVA tests are reported to compare pairs of conditions. (B) Transwell invasion assay with OptoControl iRFP-Abi1 wild-type, OptoRal iRFP-Abi1 wild-type, and OptoRal iRFP-Abi1Q56A cells, in the dark or under light stimulation. Graph shows the mean ±SEM from five independent experiments. In the graph the results of one-way ANOVA tests are reported to compare pairs of conditions. *p<0.05, **p<0.01, ***, p<0.001, ns not-significant.
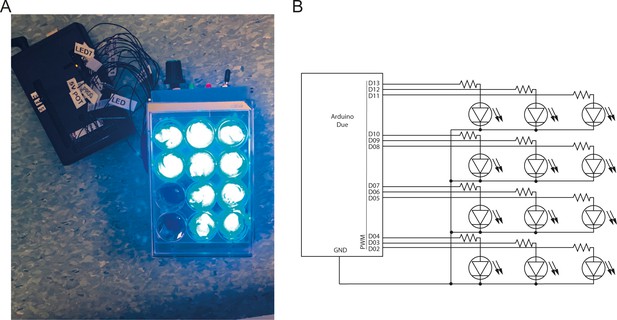
The custom-made illumination device for optogenetic activation on multi-well dishes.
(A) The device consists of an array of 12 blue LEDs mounted in a 3D-printed enclosure so that each LED illuminates one well only of a 12-well plate. Cells were illuminated from the bottom of the plate and the device was kept in a cell-culture incubator for the whole duration of the experiments. (B) Each LED is controlled independently through an Arduino Due micro-controller.
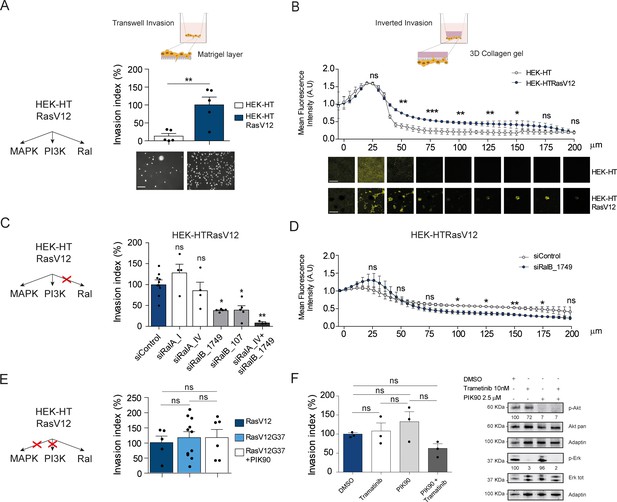
Ras-driven invasion requires RalB, rather than MAPK and PI3K.
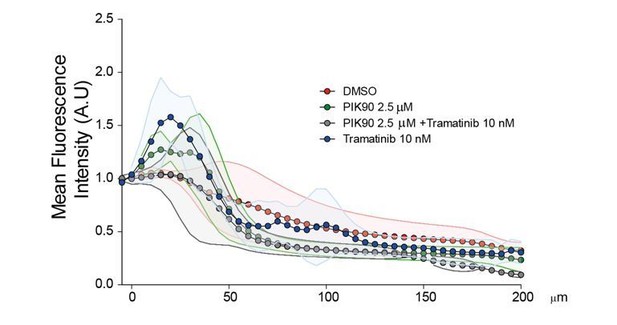
(A) Comparison of invasive capability of HEK-HT and HEK-HT-H-RasV12 cells through a thin matrigel layer using Transwell invasion assay (6 hr). Representative images of invading cells stained with DAPI are shown below the graph. Scale bar, 50 µm. Graph shows the mean ±SEM. Each dot represents one experiment. For statistics Student t-test was used. (B) Comparison of invasive capability of HEK-HT and HEK-HT-HRasV12 cells through a 3D bovine collagen gel (2.3 mg/ml) using Inverted invasion assay (4 days). Cells were live stained with CellTrace. Invasion was quantified as mean fluorescence of confocal fields acquired every 5 µm starting from below the porous membrane till 200 µm distance inside the gel. Graph shows the mean ±SD of measurements in duplicate from three experiments. Representative images are shown below the graph. Scale bar, 250 µm. Statistical comparison of the curves was done using Student t-test at every 25 µm. (C) RalB but not RalA is required for HEK-HTRasV12 invasion through a matrigel layer (Transwell invasion assay). Cells were transfected with the indicated siRNAs. For statistics one-way ANOVA test was used. (D) RalB is required for HEK-HTRasV12 invasion through a 3D collagen gel (Inverted invasion assay). Invasion was quantified as in panel B. (E) Comparison of invasive capability of HEK-HT-H-RasV12, HEK-HT-H-RasV12G37 and PIK90-treated HEK-HT-H-RasV12G37 cell lines using Transwell invasion assay. The combination of the additional G37 mutation and treatment with the PI3K inhibitor PIK90 (10 µM, treatment started 1 hr before Transwell invasion assay and maintained during the 6 hr invasion) impairs RasV12-dependent MAPK and PI3K hyperactivation, conserving only the activation of Ral pathway. For statistics one-way ANOVA test was used. (F) Comparison of invasive capability of HEK-HT-H-RasV12 cells treated with DMSO, with the MEK inhibitor Trametinib (10 nM), with the PI3K inhibitor PIK90 (2.5 μM), or with both Trametinib and PIK90. Treatments were started 1 hr before Transwell invasion assay and maintained during the 6 hr invasion. Cell lysates were prepared at 1 hr post-treatment. On the right, representative western blots for total Akt and phospho-Akt (Ser437), and for total Erk and phospho-Erk (Thr202/Tyr204), show the efficient pharmacological blockage of MAPK and PI3K pathways. Quantifications of Akt and Erk phosphorylation, calculated as p-Akt/total Akt or p-Erk/total Erk ratios, and normalized for HEK-HT-HRasV12 without drugs, are shown below the WBs. For statistics one-way ANOVA test was used. *p<0.05, **p<0.01, ***p<0.001, ns not-significant.
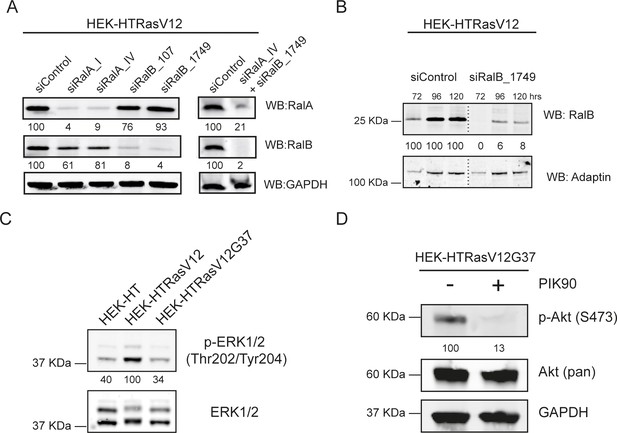
Representative western blots.
(A) Validation of Ral depletions for Transwell invasion assay. Representative western blots for RalA and RalB from cell lysates of HEK-HT-RasV12 cells, prepared 72 hr after transfection with the indicated siRNA. GAPDH is used as loading control. Quantifications of Ral depletion efficiencies, calculated as Ral/GAPDH ratio and normalized for siControl condition, are shown below the WBs. (B) Validation of Ral depletions for Inverted invasion assay. Representative western blots for RalA and RalB from cell lysates of HEK-HT-RasV12 cells, prepared 72 hr, 96 hr and 120 hr after transfection with the indicated siRNA. Adaptin is used as loading control. Quantifications of Ral depletion efficiencies, calculated as Ral/adaptin ratio and normalized for siControl condition at the same time point, are shown below the WBs. (C) Inhibition of MAPK pathway by G37 mutation. Representative western blot for total Erk and phospho-Erk (Thr202/Tyr204) from cell lysates of HEK-HT, HEK-HT-HRasV12, and HEK-HT-HRasV12G37 cells. Quantifications of Erk phosphorylation, calculated as p-Erk/total Erk ratio and normalized for HEK-HT-H-RasV12, are shown below the WBs. (D) Inhibition of PI3K pathway by PIK90. PIK90 (10 µM) treatment was started 1 hr before Transwell invasion assay and maintained during the 6 hr invasion. Cell lysates were prepared at 1 hr post-treatment. Representative western blot for total Akt and p-Akt (Ser437) from cell lysates of HEK-HT-HRasV12G37 cells, treated or not with PIK90 (10 µM). GAPDH is used as loading control. Quantifications of Akt phosphorylation, calculated as p-Akt/total Akt ratio and normalized for HEK-HT-H-RasV12G37 without PIK90, are shown below the WBs.
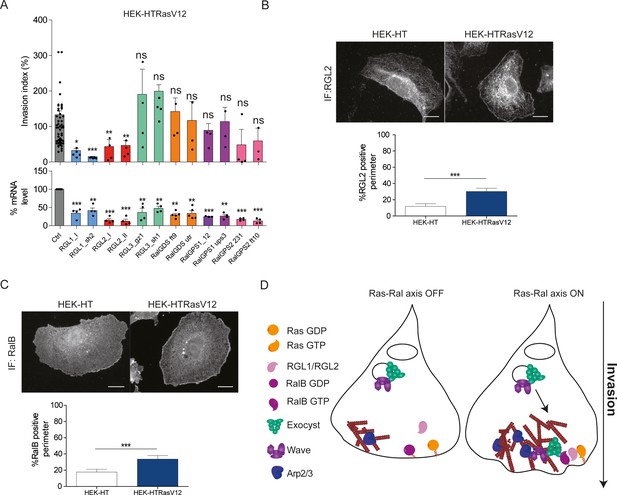
Ras-driven invasion requires the two RalGEF RGL1 and RGL2.
(A) HEK-HTRasV12 cells require RGL1 and RGL2 for Transwell invasion (upper panel). Depletions of the six RalGEF were validated by RT-qPCR (lower panel). Graphs show the mean ±SEM. Each dot represents one experiment. For statistics one-way ANOVA was used. *p<0.05, **p<0.01, ***p<0.001, ns not-significant. (B–C) Localization of endogenous RGL2 and RalB is increased at the cell edges of HEK-HTRasV12 transformed cells as compared to HEK-HT. Representative epifluorescence images of the immunostaining of RGL2 and RalB are depicted. Scale bar, 10 µm. Graphs show the averaged percentage ±SEM of cell perimeter which is positive for RGL2 or RalB. n > 40 cells from four independent experiments. For statistics Student t-test was used. *p<0.05, **p<0.01, ***, p<0.001, ns indicates not-significant. (D) The Ras-RGL1/2-RalB-exocyst-WRC axis drives invasion. Active Ras-GTP binds and recruit the two RalGEFs RGL1 and RGL2, which activate RalB; RalB-GTP binds to the exocyst complex, promoting its assembly and recruitment to the leading edge; by its direct association with the WRC complex, exocyst drives WRC to the leading edge, where WRC stimulates actin polymerization via the Arp2/3 complex, consequently promoting protrusion formation, motility and invasion.
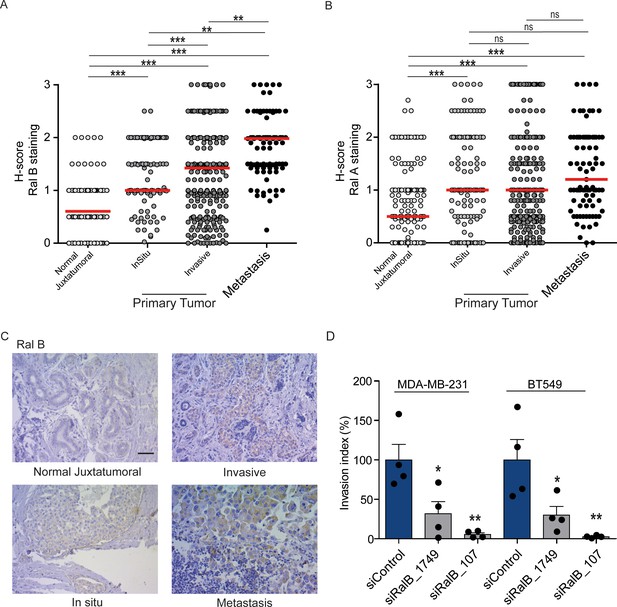
Expression of Ral proteins in human breast cancers. Role of RalB in breast cancer invasion.
(A) Quantitative analysis of RalB protein level in patient samples of normal juxtatumural tissue (n = 298), in situ carcinoma (n = 101), invasive ductal carcinoma (n = 439) and lymph node metastasis (n = 91). Since none of the data sets passed the Shapiro-Wilk normality test, the median H-scores are displayed. Wilcoxon signed-rank tests were performed on paired (i.e. from same patient) data points: n = 73 for normal versus in situ, n = 290 for normal versus invasive, n = 63 for normal versus metastasis, n = 99 for in situ versus invasive, n = 29 for in situ versus metastasis, n = 85 for invasive versus metastasis. *p<0.05, **p<0.01, ***p<0.001, ns not-significant. (B) Quantitative analysis of RalA protein level in patient samples of normal juxtatumural tissue (n = 371), in situ carcinoma (n = 144), invasive ductal carcinoma (n = 462) and lymph node metastasis (n = 87). Since none of the data sets passed the Shapiro-Wilk normality test, the median H-scores are displayed. Wilcoxon signed-rank tests were performed on paired (i.e. from same patient) data points: n = 129 for normal versus in situ, n = 368 for normal versus invasive, n = 69 for normal versus metastasis, n = 142 for in situ versus invasive, n = 39 for in situ versus metastasis, n = 83 for invasive versus metastasis. *p<0.05, **p<0.01, ***p<0.001, ns not-significant. (C) Representative RalB immunostaining in the different breast cancer compartments. Scale bar, 250 µm. (D) Invasion of human breast cancer cells requires RalB, regardless of Ras mutational status. MDA-MB-231 cells (carrying the KRasG13D mutation) and BT549 cells (Ras wild-type) were transfected with the indicated siRNAs and subjected to Transwell invasion assay. Graph shows the mean ±SEM. Each dot represents one experiment. For statistics one-way ANOVA test was used. *p<0.05, **p<0.01, ***p<0.001. ns not-significant. Legends for figure supplements.
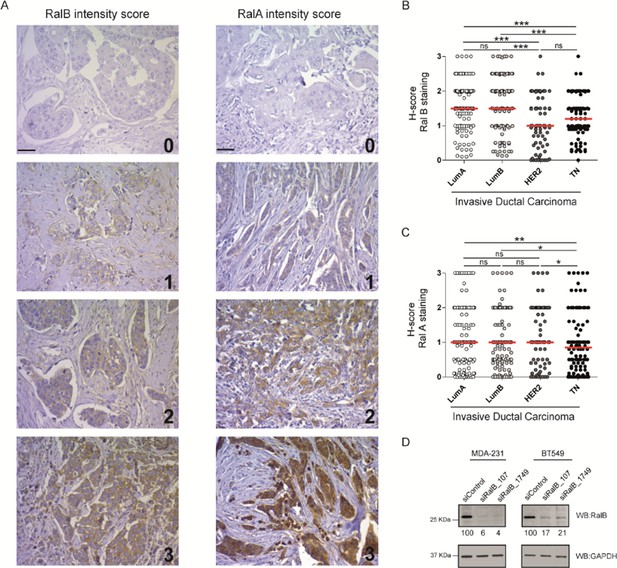
Ral proteins and breast cancers.
(A) Representative staining scores for RalB and RalA on human invasive breast carcinoma. Ral proteins are stained brown (DiAminoBenzidine) and cell nuclei are stained blue (hematoxylin). Scale bar, 250 µm. (B) RalB protein level in invasive ductal carcinoma, according to the molecular breast cancer subtypes classification. Since none of the data sets passed the Shapiro-Wilk normality test, the median H-scores are displayed. Mann-Whitney statistical tests were performed. *p<0.05, **p<0.01, ***p<0.001, ns not-significant. (C) RalA protein level in invasive ductal carcinoma, according to the molecular breast cancer subtypes classification. Since none of the data sets passed the Shapiro-Wilk normality test, the median H-scores are displayed. Mann-Whitney statistical tests were performed. *p<0.05, **p<0.01, ***p<0.001, ns not-significant. (D) Validation of RalB depletions in MDA-MB-231 and BT549 cells. Representative western blots from cell lysates of cells treated with the indicated siRNA. GAPDH is used as loading control. Quantifications of RalB depletion efficiencies, calculated as Ral/GAPDH ratio and normalized for siControl condition, are shown below the WBs.
Videos
Blue light induces local recruitment at plasma membrane of RalGEF-CRY2-mCherry in OptoRal cells.
OptoRal cells were imaged by TIRF microscopy (Cherry channel). Cells were illuminated inside the blue rectangle starting at time t = 2.15 min. Scale bar is 10 μm. Time length: 8.5 min. Images were acquired every 15 s and the video is shown at 7 frame/s.
Blue light induces local Ral activation in OptoRal cells.
OptoRal cells expressing RalB-GFP and Sec5GBD-iRFP were imaged by TIRF microscopy. On the left, the recruitment of RalGEF is followed in the mCherry channel. On the right, the recruitment of Sec5GBD (reporter for Ral activity) is followed in the iRFP channel. Cells were illuminated inside the blue rectangle starting at time t = 2 min. Scale bar is 10 μm. Time length: 32.25 min. Images were acquired every 15 s and the video is shown at 7 frame/s.
Local activation of Ral induces protrusions.
On the left, optogenetic recruitment of of RalGEF(Cherry channel) in OptoRal cells imaged by TIRF microscopy. On the right, threshold-based binary mask of the cells used to calculate the edge velocity. Cells were illuminated inside the blue rectangle starting at time t = 3.10 min. Scale bar is 10 μm. Time length: 6.83 min. Images were acquired every 10 s and the video is shown at 7 frame/s.
Light stimulation of OptoControl cells does not induce protrusions.
Same conditions as Video 3.
Local activation of Ral induces WRC recruitment at plasma membrane.
OptoRal cells expressing iRFP-Abi WT were imaged by TIRF microscopy. On the left, the recruitment of RalGEF is followed in the mCherry channel. On the right, the recruitment of Abi1 (a subunit of WRC complex) is followed in the iRFP channel. Cells were illuminated inside the blue rectangle starting at t = 5 min. Scale bar is 10 μm. Time length: 10.17 min. Images were acquired every 10 s and the videos are shown at 7 frame/s.
Light stimulation of OptoControl cells does not induce WRC recruitment at plasma membrane.
Same conditions as Video 5.
Additional files
-
Supplementary file 1
List of cell lines
- https://doi.org/10.7554/eLife.40474.023
-
Supplementary file 2
List of plasmids
- https://doi.org/10.7554/eLife.40474.024
-
Supplementary file 3
List of siRNAs
- https://doi.org/10.7554/eLife.40474.025
-
Supplementary file 4
List of primers
- https://doi.org/10.7554/eLife.40474.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40474.027





