Structures of translationally inactive mammalian ribosomes
Figures
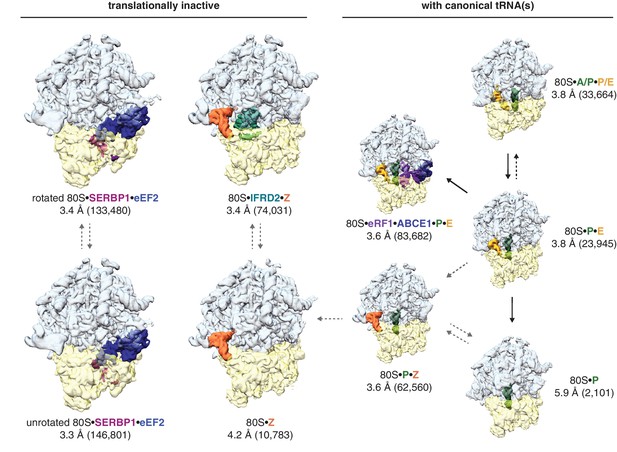
Identification of translationally inactive ribosomal complexes in a cryo-EM dataset.
In silico classification of a cryo-EM dataset of ribosomal particles reveals known intermediates of translation and translationally inactive 80S ribosomes. The overall resolution and number of particles that make up the reconstruction are shown for each complex.
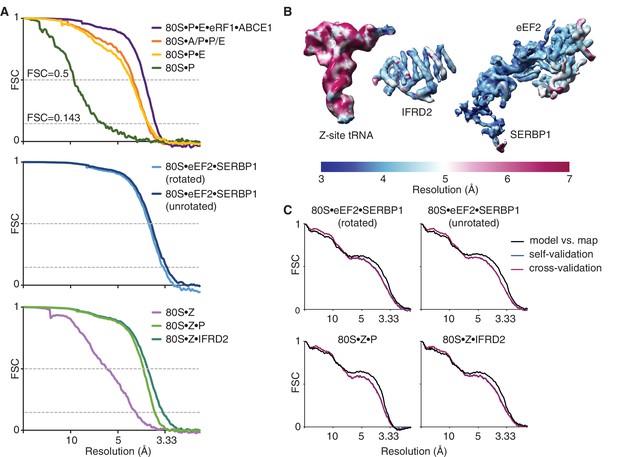
Global and local resolution.
(A) Fourier-shell-correlation (FSC) curves for each reported cryo-EM reconstruction. FSC values at 0.5 and 0.143 are indicated with dashed lines. (B) The cryo-EM map for each factor colored by local resolution. (C) Validation curves.
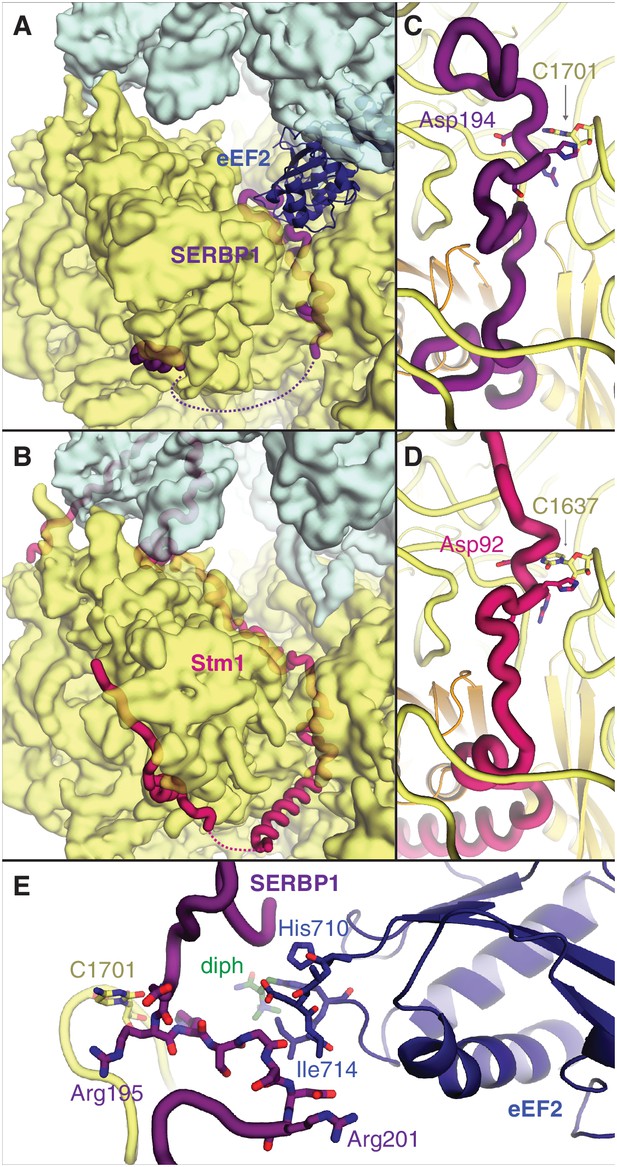
SERBP1 sequesters eEF2 on inactive ribosomes.
Comparison of (A) SERBP1 (SERPINE1 mRNA-binding protein 1) and eEF2 (eukaryotic elongation factor 2) bound to the rabbit ribosome with (B) Stm1 bound to the yeast ribosome (PDB 4V88). (C) Path of SERBP1 and (D) Stm1 through the mRNA entrance channel. (E) Interaction between SERBP1 and eEF2.
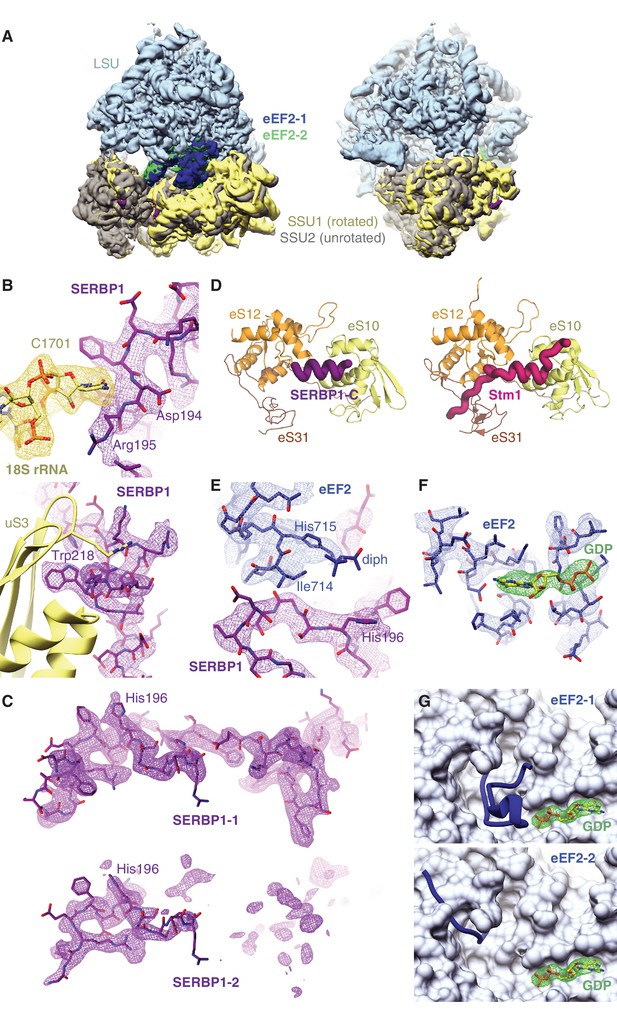
Distinct classes of eEF2 and SERBP1 bound to 80S ribosomes.
(A) Overview of the two classes of 80S ribosomes bound to eEF2 and SERBP1 (see Figure 1). The small ribosomal subunit (SSU) in the rotated conformation is colored in yellow, with the corresponding eEF2 density colored in blue. The other unrotated class is aligned to this map via the large ribosomal subunit (LSU), with the corresponding SSU and eEF2 densities colored in grey and green, respectively. (B) Density of SERBP1 (purple) displaying interactions with 18S rRNA and the uS3 protein in the mRNA entrance channel of the small ribosomal subunit (yellow). (C) Comparison of the EM map density for SERBP1 (purple) in the rotated (top) or unrotated (bottom) ribosome. (D) Comparison of the interactions between the C-terminus of either rabbit SERBP1 (purple, left panel) or yeast Stm1 (pink, right panel) with the indicated proteins of the small ribosomal subunit. (E) Map density of the interaction between eEF2 (blue) and SERBP1 (purple). The diphthamide (diph) modification on His715 of eEF2 is indicated. (F) Map density for the GDP nucleotide (green) bound to eEF2 (blue). (G) Comparison of different conformations of the eEF2 nucleotide-binding domain on the rotated (top) and unrotated (bottom) ribosome.

Sequence conservation of Stm1 and SERBP1.
Sequence alignment of four isoforms of rabbit SERBP1 and yeast Stm1, with highly conserved residues highlighted in blue. The segments of SERBP1 isoform two modeled from this work are shown above the sequence alignment, depicted as secondary structure.
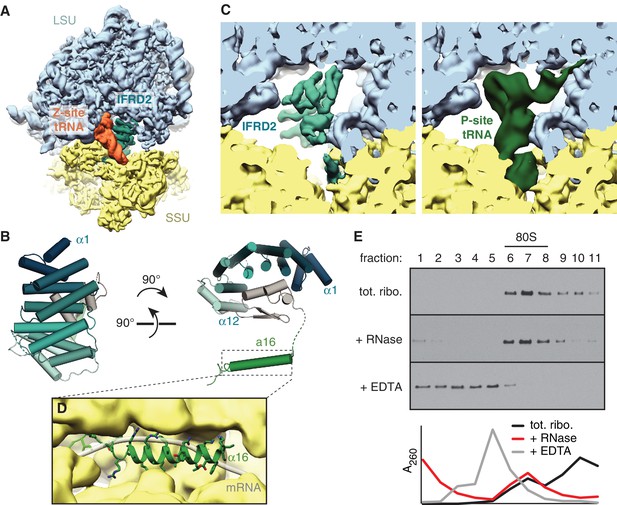
IFRD2 binding to ribosomes is incompatible with translation.
(A) IFRD2 (interferon-related developmental regulator 2) occupies the intersubunit space in a subset of ribosomes containing Z-site tRNA. (B) Model of IFRD2 colored by domain. (C) IFRD2 interacts with many of the same ribosomal elements as P-site tRNA. (D) The C-terminal α-helix of IFRD2 occupies the mRNA exit channel and follows the path taken by mRNA through the ribosome. (E) 200 nM ribosomes isolated from rabbit reticulocyte lysate were treated without or with 50 μg/mL RNase A or 10 mM EDTA for 5 min and subjected to native size fractionation on 10 – 50% sucrose gradients. Eleven fractions taken from the top were collected and subjected to immunoblotting for IFRD2. Absorbance readings at 254 nm are shown below.
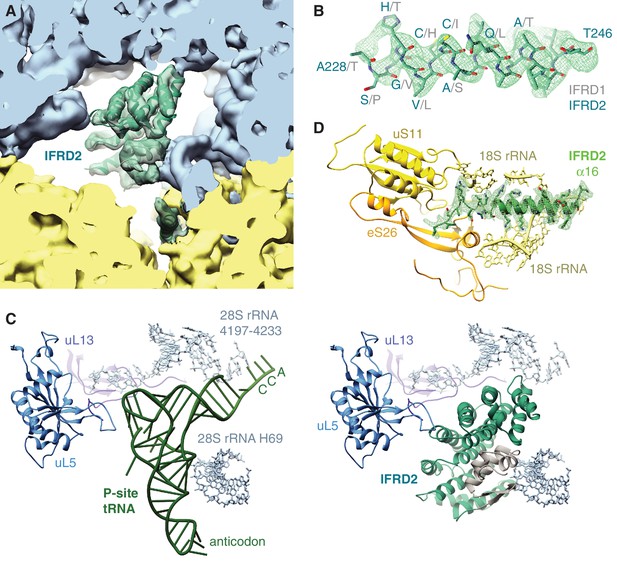
IFRD2 interactions with 80S ribosomes.
(A) Fit of the IFRD2 model (teal) to the EM density map. (B) EM density corresponding to helix 8 of IFRD2, with corresponding residues of IFRD2 and aligned residues of IFRD1 (grey) indicated. (C) Comparison of interactions of P-site tRNA (green, left panel) and IFRD2 (right panel) with rRNA and proteins (uL13 and uL5) of the large ribosomal subunit. (D) Density of the C-terminal tail of IFRD2 (green), displaying interactions with 18S rRNA, uS11, and eS26 in the mRNA exit channel.

Sequence alignment of IFRD family members of selected eukaryotes.
Alignment of IFRD family paralog sequences from multiple eukaryotes, with highly conserved residues highlighted in purple. Secondary structure corresponding to the atomic model of IFRD2, colored as in Figure 3, is shown above.
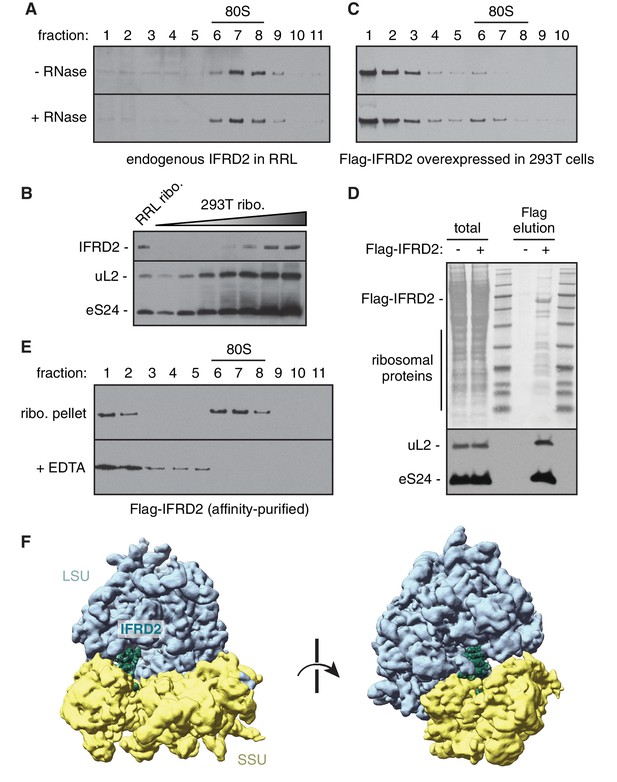
Characteristics of IFRD2 association with mammalian ribosomes.
(A) Rabbit reticulocyte lysate was directly treated without or with 50 μg/mL RNase A for 5 min and subjected to size fractionation on a 10 – 50% sucrose gradient. Eleven fractions collected from the top of the gradient were analyzed by SDS-PAGE and immunoblotting for IFRD2. Migration of 80S ribosomes is indicated. Note that all detectable endogenous IFRD2 is in ribosomal fractions. (B) Ribosomes isolated from reticulocyte lysate and HEK293T cell lysate by centrifugation were directly analyzed by SDS-PAGE and immunoblotting for the indicated ribosomal proteins and IFRD2. Comparing 0.2 pmol of reticulocyte ribosomes (lane 1) with a serial twofold titration of HEK293T ribosomes reveals an enrichment of IFRD2 on the former. (C) HEK293T cells overexpressing Flag-tagged IFRD2 were lysed and directly analyzed as in (A) and immunoblotting against the Flag epitope. (D) Lysates from HEK293T cells overexpressing Flag-tagged IFRD2 were subject to affinity purification via the Flag epitope. The elution and 10% equivalent input were analyzed by SDS-PAGE, Ponceau staining (top) and immunoblotting for ribosomal proteins, revealing that IFRD2 copurifies with ribosomes. (E) Ribosomes (ribo.) in the elution in (D) were pelleted and subjected to native size fractionation on a 10–50% gradient without or with 10 mM EDTA. Eleven fractions from the top were collected and analyzed by SDS-PAGE and immunoblotting for Flag-tagged IFRD2. (F) Cryo-EM map of Flag-tagged IFRD2 on human 80S ribosomes, with the model of IFRD2 (Figure 3) docked.
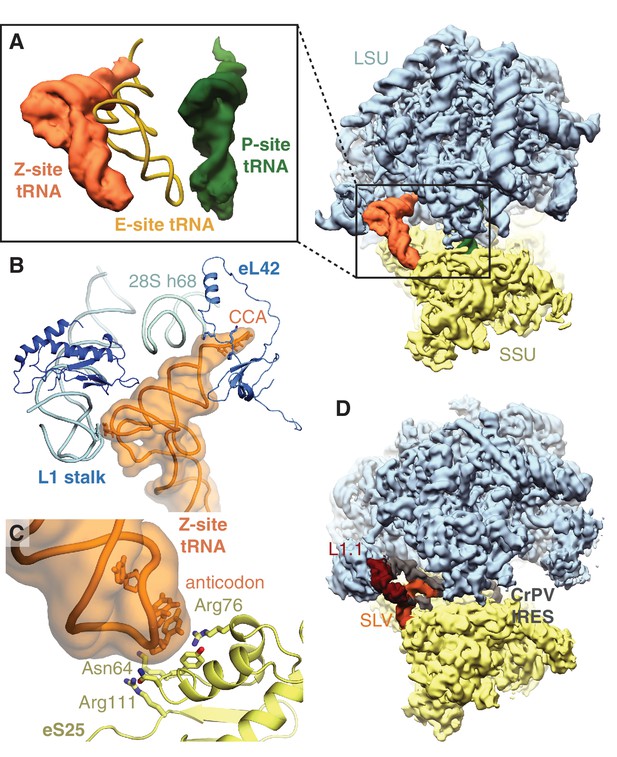
Z-site tRNA.
(A) Position of Z-site tRNA on the ribosome in the presence of a P-site tRNA. Docked E-site tRNA (shown in cartoon representation) is incompatible with a Z-site tRNA. (B) Interactions between the Z-site tRNA (CCA nucleotides shown in stick representation) and the 60S ribosomal subunit. (C) Interaction between the anticodon stem-loop (anticodon nucleotides shown in stick representation) of Z-site tRNA and eS25. (D) Cryo-EM map of CrPV IRES (EMD-2599), with the L1.1 and stem-loop V (SLV) domains, which make similar interactions as Z-site tRNA, indicated, bound to the Kluyveromyces lactis ribosome. LSU – large ribosomal subunit; SSU – small ribosomal subunit; IRES – internal ribosome entry site; SLV – stem-loop V.
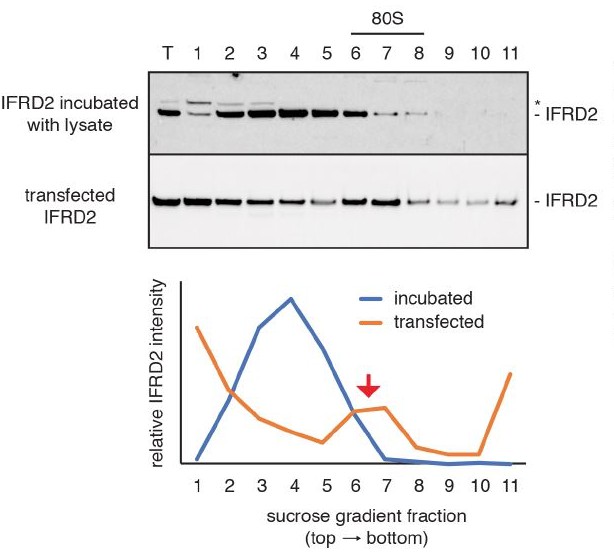
Assembly of IFRD2 onto ribosomes.
Recombinant IFRD2 purified from E. coli was incubated with cytosol isolated from HEK293T cells for 30 minutes at 32°C and subject to native size fractionation by centrifugation on a 10-50% sucrose gradient. Eleven fractions were collected from the top of the gradient and analyzed by SDS-PAGE and immunoblotting for the recombinant IFRD2 (top blot; quantified by blue curve). This revealed that only minor co-association of IFRD2 in ribosomal (80S) fractions. In contrast, transiently transfecting IFRD2 into HEK293T cells, followed by cytosol isolation and size fractionation (bottom plot; quantified by orange curve) reveals a distinct peak of IFRD2 co-association in ribosomal fractions (red arrow).
Tables
Structurally distinct classes identified within a cryo-EM dataset of 971,191 ribosomal particles.
https://doi.org/10.7554/eLife.40486.011| Ribosomal complex | Resolution (Å) | Particles | % of particles | EMDB accession code |
|---|---|---|---|---|
| 80S classes (unrotated) | ||||
| 80S • P | 5.9 | 2101 | 0.2 | 9234 |
| 80S • P • E | 3.8 | 23,945 | 2.5 | 9235 |
| 80S • P • E • eRF1 • ABCE1 | 3.6 | 83,682 | 8.6 | - |
| 80S • Z | 4.2 | 10,783 | 1.1 | 9236 |
| 80S • Z • P | 3.6 | 62,560 | 6.4 | 9237 |
| 80S • Z • IFRD2 | 3.4 | 74,031 | 7.6 | 9239 |
| 80S • eEF2 • SERBP1 (head swivel) | 3.3 | 146,801 | 15.1 | 9240 |
| 80S classes (rotated) | ||||
| 80S • A/P • P/E | 3.8 | 33,664 | 3.5 | 9241 |
| 80S • eEF2 • SERBP1 | 3.4 | 133,480 | 13.7 | 9242 |
Model statistics.
https://doi.org/10.7554/eLife.40486.013| 80S • Z • P | 80S • Z • IFRD2 | 80S • eEF2 • SERBP1 (head swivel) | 80S • eEF2 • SERBP1 (rotated) | |
|---|---|---|---|---|
| Model composition | ||||
| Protein residues | 11,547 | 11,901 | 12,760 | 12,591 |
| RNA bases (+modified bases) | 5686 (40) | 5603 (26) | 5534 (36) | 5528 (40) |
| Ligands (Zn2+/Mg2+) | 8/297 | 8/311 | 8/297 | 8/298 |
| Refinement | ||||
| Resolution (Å) | 3.6 | 3.4 | 3.3 | 3.4 |
| Map sharpening B factor (Å2) | −101.9 | −95.5 | −97.1 | −95.1 |
| Average B factor (Å2) | 75.6 | 51.8 | 84.1 | 86.1 |
| Correlation coefficient volume mask | 0.844 0.865 | 0.825 0.853 | 0.847 0.862 | 0.847 0.862 |
| Rms deviations | ||||
| Bond lengths (Å) | 0.010 | 0.009 | 0.007 | 0.008 |
| Bond angles (°) | 1.2 | 1.1 | 1.1 | 1.0 |
| Validation (proteins) | ||||
| MolProbity score | 1.79 | 1.64 | 1.68 | 1.71 |
| Clashscore, all atoms | 5.8 | 5.0 | 5.6 | 5.7 |
| Rotamer outliers (%) | 1.3 | 0.8 | 0.9 | 0.9 |
| EMRinger score | 2.87 | 3.24 | 2.86 | 2.97 |
| Ramachandran plot | ||||
| Favored (%) | 94.0 | 94.3 | 94.5 | 94.0 |
| Outliers (%) | 0.1 | 0.1 | 0.1 | 0.1 |
| Validation (RNA) | ||||
| Probably wrong sugar puckers (%) | 1.6 | 1.5 | 1.6 | 1.4 |
| Bad backbone conformations (%) | 23.6 | 23.6 | 22.8 | 22.0 |
| PDB | 6MTB | 6MTC | 6MTD | 6MTE |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell Line (H. sapiens) | HEK293T | American Type Culture Collection | Cat# CRL-3216 RRID:CVCL_0063 | |
| Recombinant DNA reagent | pcDNA3.1-3xFLAG-TEV-IFRD2 | This paper | N/A | Mammalian expression vector expressing Flag-tagged IFRD2 behind a CMV promoter |
| Antibody | IFRD2 | Invitrogen | Cat# PA5-48833 RRID:AB_2634289 | IB: 1:1000 |
| Antibody | FLAG M2 | Sigma-Aldrich | Cat# F3165 RRID:AB_259529 | IB: 1:5000 |
| Antibody | uL2 | Abcam | Cat# ab169538 RRID:AB_2714187 | IB: 1:10000 |
| Antibody | eS24 | Abcam | Cat# ab196652 RRID:AB)2714188 | IB: 1:3000 |
| Antibody | HRP anti-mouse | Jackson ImmunoResearch | Cat# 115-035-003 RRID:AB_10015289 | IB: 1:5000 |
| Antibody | HRP anti-rabbit | Jackson ImmunoResearch | Cat# 111-035-003 RRID:AB_2313567 | IB: 1:5000 |
| Antibody | FLAG M2 agarose resin | Sigma-Aldrich | Cat# A2220 RRID:AB_10063035 | |
| Peptide, recombinant protein | 3X FLAG peptide | Sigma-Aldrich | Cat# F4799 | |
| Peptide, recombinant protein | RNase A | Sigma-Aldrich | Cat# R6513 | |
| Chemical compound, drug | Dulbecco's Modified Eagle Medium (DMEM) | Gibco/Thermo Fisher | Cat# 10569 | |
| Chemical compound, drug | HyClone Fetal Bovine Serum | GE Healthcare Life Sciences | Cat# SH30910.03 | |
| Chemical compound, drug | Trypsin-EDTA | Gibco/Thermo Fisher | Cat# 25200 | |
| Chemical compound, drug | OptiMEM | Gibco/Thermo Fisher | Cat# 31985 | |
| Chemical compound, drug | TransIT 293 | Mirus | Cat# MIR2705 | |
| Chemical compound, drug | EDTA | GrowCells | Cat# MRGF-1202 | |
| Chemical compound, drug | SuperSignal West Pico | Thermo Fisher | Cat# 34080 | |
| Biological sample (O. cuniculus) | Rabbit reticulocyte lysate | Green Hectares | N/A | |
| Software, algorithm | RELION-2.0 or RELION-2.1 | Kimanius et al., 2016 | RRID:SCR_016274 | https://www2.mrc-lmb.cam.ac.uk/relion |
| Software, algorithm | MotionCor2 | Zheng et al., 2017 | RRID:SCR_016499 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Software, algorithm | GCTF | Zhang, 2016 | RRID:SCR_016500 | https://www.mrc-lmb.cam.ac.uk/kzhang/ |
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera/ |
| Software, algorithm | Coot v.0.8.9 | Brown et al., 2015a | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software, algorithm | I-TASSER | Zhang, 2008 | RRID:SCR_014627 | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ |
| Software, algorithm | PHENIX | Adams et al., 2010 | RRID:SCR_014224 | https://www.phenix-online.org/ |
| Software, algorithm | MolProbity v.4.3.1 | Chen et al., 2010 | RRID:SCR_014226 | http://molprobity.biochem.duke.edu/ |
| Software, algorithm | EMRinger | Barad et al., 2015 | N/A | http://emringer.com/ |
| Software, algorithm | SBGrid | Morin et al., 2013 | RRID:SCR_003511 | https://sbgrid.org/ |
| Software, algorithm | PyMOL | DeLano, 2002 | RRID:SCR_000305 | http://www.pymol.org |
| Software, algorithm | SCIPION | de la Rosa-Trevín et al., 2016 | N/A | http://scipion.i2pc.es/ |
| Software, algorithm | MonoRes | Vilas et al., 2018 | N/A |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40486.014
-
Supplementary file 1
Curated list of the proteins observed by mass spectrometry used to identify factors in the cryo-EM maps.
For clarity, ribosomal proteins and contaminating bacterial and skin proteins are excluded and known components of multisubunit complexes clustered at the end of the table. Sample A represents the sample used for cryo-EM. Sample B are the proteins eluted from ribosomes under physiological salt conditions. Sample C are the proteins eluted from ribosomes under high-salt (750 mM KOAc, 15 mM Mg(OAc)2) conditions. Proteins observed in cryo-EM complexes are highlighted in yellow.
- https://doi.org/10.7554/eLife.40486.015





