Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis
Figures
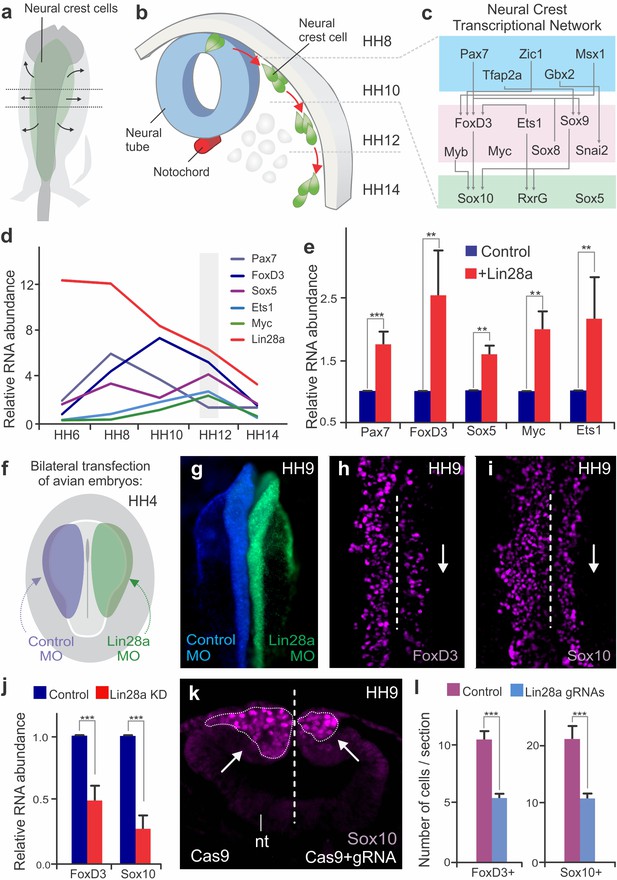
Changes in Lin28a levels impact neural crest development in vivo.
(a–b) Neural crest migration during avian development. (a) Neural crest progenitor cells (green) are specified on dorsal folds of the neural tube (grey) during early development. (b) Transverse section of the neural tube showing the position of neural crest cells through development, as they progressively move away from the neural tube to differentiate. HH8 and HH14 are the earliest and latest developmental stages shown in the diagram, respectively. (c) A schematic of the early gene regulatory network composed of transcription factors involved in neural crest cells formation. (d) Expression levels of Lin28a and transcription factors of the early gene regulatory circuit, in sorted neural crest cells obtained from different stages. (e) Constitutive expression of Lin28a results in maintenance of multipotency genes in late neural crest cells. RT-PCR for Pax7, FoxD3, Sox5, Myc and Ets1 comparing the expression of these genes in control vs. Lin28a overexpressing migratory neural crest cells. (f) Electroporation scheme for loss-of-function assays in which control reagent (blue) and targeted reagent (green) were injected in different sides of a HH4 chick embryo. (g) Dorsal whole mount view of HH9 embryo with Control MO on the left and Lin28a MO on the right. Immunohistochemistry for neural crest markers FoxD3 (h) and Sox10 (i) on Lin28a knockdown. Dotted line represents embryo midline (j) RT-PCR for FoxD3 and Sox10 transcripts in control vs Lin28a MO treated neural folds. (k–l) CRISPR-Cas9 mediated knockdown of Lin28a recapitulates the MO phenotype. (k) Transverse section showing Sox10 positive cells in control and knockdown sides of the embryo head, showing reduction in the number of neural crest cells (arrow). (l) Quantification of FoxD3+ and Sox10+ cells following CRISPR-Cas9 mediated knockdown of Lin28a. Error bars in (e), (j) and (l) represent standard error. HH: Hamburger and Hamilton developmental stages, MO: Morpholino.
-
Figure 1—source data 1
The RT-PCR results of Lin28a over-expression and morpholino-mediated knockdown experiments and the quantitation of FoxD3 +and Sox10 +cells following CRISPR.
- https://doi.org/10.7554/eLife.40556.007
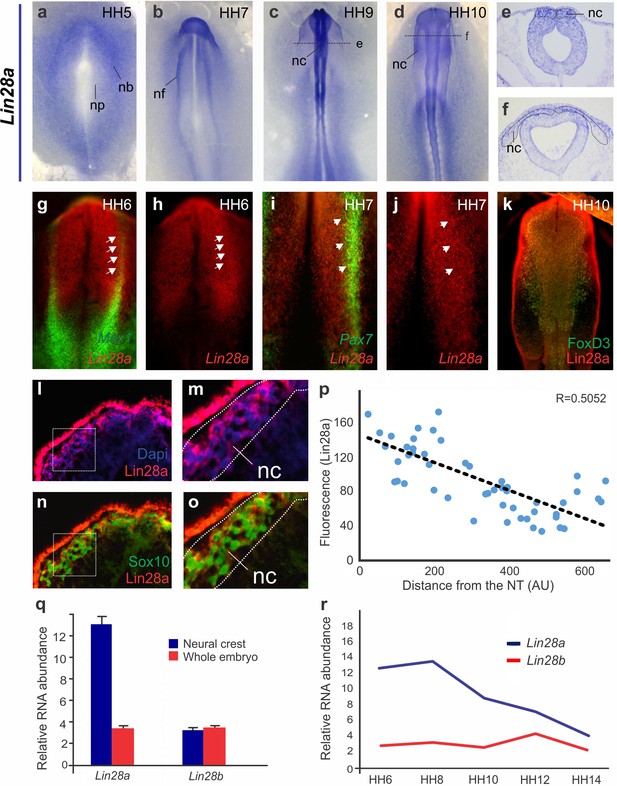
Expression patterns of Lin28a and Lin28b mRNA and Lin28a protein during early chick development.
(a–f) Colorimetric in situ hybridization for Lin28a in chick embryos of different developmental stages. Lin28a mRNA is enriched in the neural plate border at HH5 (a), in the dorsal neural folds at stage HH7-9 (b–c) and in migrating neural crest at stage HH10 (d). Transverse sections showing Lin28a expression in pre-migratory and migratory neural crest cells (e–f). (g–j) Fluorescent in situ hybridization for Lin28a and early neural crest genes Msx1 and Pax7. At HH6, Lin28a expression overlaps with Msx1 (g–h) and at HH7 with Pax7 in the neural plate border (arrowheads) (i–j). (k) Immunohistochemistry for Lin28a protein, and neural crest markers FoxD3. In HH10 embryos, Lin28a protein (red) is expressed in FoxD3+ (green) neural crest cells (l–o). Transverse sections showing the localization of the Lin28a protein in the cytoplasm (l–m) of Sox10 +migratory neural crest cells (n–o). (p) Quantification of Lin28a fluorescence in migratory neural crest cells, showing that levels of Lin28a protein decrease as cells migrate away from the neural tube. (q) RT-PCR for Lin28a and Lin28b in FACS sorted neural crest (NC) cells and in whole embryo (WE) at HH8, showed that Lin28a, but not Lin28b, is significantly enriched in neural crest cells. (r) Relative expression levels of Lin28b paralog (red line) in FACS sorted neural crest cells at different developmental stages highlight that Lin28b is lowly expressed in neural crest cells and does not recapitulate the expression dynamics of Lin28a. The expression level of Lin28a (blue line) at the same developmental timepoints, shown in Figure 1, has been included here for comparison. AU: arbitrary units. np: Neural plate, nb: neural plate border, nf: neural fold, nc: neural crest, nt: neural tube.
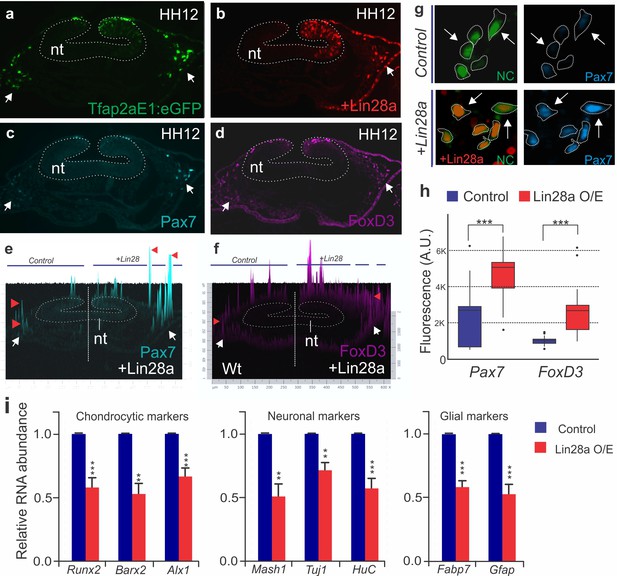
Ectopic expression of Lin28a prevents silencing of early neural crest genes and delays differentiation.
Representative transverse section of an HH12 embryo, bilaterally injected with Tfap2aE1:eGFP on the left, and with Tfap2aE1:eGFP and pCI:Lin28a-H2B-RFP on the right. GFP expressing neural crest cells are shown in green (arrows) (a) and cells expressing Lin28a-H2B-RFP are shown in red (b). Immunostaining for Pax7(c) and FoxD3 (d) revealed a sustained expression of these proteins in the migratory neural crest on cells transfected with the Lin28a expression vector (arrows, e–f). 2.5D fluorescence intensity plots show that Pax7(e) and FoxD3 (f) intensity is higher in neural crest cells on the right side of the embryo, which express Lin28a in high levels. The height of the peaks (red arrowheads) is a measure of the fluorescence intensity of the proteins in the corresponding cells. (g) Insets showing Pax7 protein expression in control neural crest cells (green) and in cells overexpressing Lin28a (red). Dotted lines mark outlines of neural crest cells. (h) Boxplots showing the fluorescence intensity of FoxD3 and Pax7 in cells from control and Lin28a overexpression side of the embryo. Nt: neural tube, HH: Hamburger and Hamilton stages. (i) RT-PCR comparing the expression of differentiation markers in control and LIN28a O/E neural crest cells from HH12 embryos, show that chondrocytic (Runx2, Barx2, and Alx1), neuronal (Mash1, Tuj1 and HuC) and glial differentiation (Fabp7 and Gfap) is delayed following sustained Lin28a expression. A.U: Arbitrary units, nt: Neural tube, O/E: over-expression.
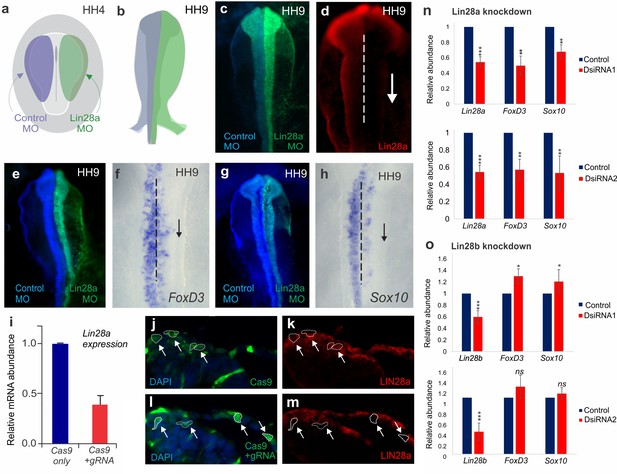
Supporting data for Lin28a and Lin28b loss-of-function analysis.
(a–b) Transfection strategy for knockdown experiments. HH4 embryos were injected with the control morpholino (blue) on the left and with the Lin28a morpholino (green) on the right (a). Following electroporation, embryos were cultured in albumin and incubated until HH9, when they were screened for efficient transfection and analyzed (b). (c) Dorsal view of a bilaterally electroporated HH9 embryo, showing control MO (left) and Lin28a MO (right). (d) Immunohistochemistry for Lin28a showed loss of the protein on the experimental side (downward arrow). (e–h) In situ hybridization for neural crest markers FoxD3 and Sox10 following knockdown of Lin28a. (i–m) Disruption of Lin28a expression with CRISPR/Cas9. (i) Quantitative analysis of Lin28a transcripts following CRISPR-cas9 targeting of the protein through RT-PCR. Immunohistochemistry for Cas9 and Lin28a on transverse sections of embryos electroporated with either Cas9 alone (j–k) or with Cas9 vector containing a pair of gRNAs targeting Lin28a. (l–m) Dotted lines outline cells expressing Cas9 (arrows). (n) RT-PCR for Lin28a, FoxD3 and Sox10, following knockdown of Lin28a using two different DsiRNAs, confirm that loss of the protein results in reduced expression of these early neural crest markers. (o) Downregulation of Lin28b using two independent DsiRNAs followed by quantification of FoxD3 and Sox10 transcripts reveal that Lin28b is not required for neural crest formation. MO: Morpholino, DsiRNA: Dicer-substrate siRNA, gRNA: guide-RNA, HH: Hamburger and Hamilton stages.
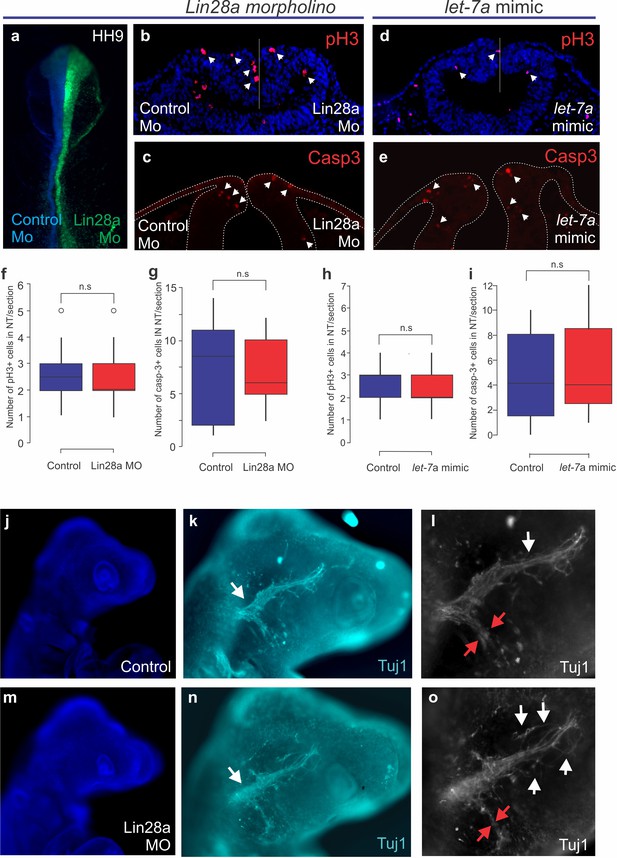
Effects of Lin28a loss-of-function on cell death, proliferation and the morphology of cranial ganglia.
(a–i) Disruption of Lin28a/let-7 axis does not cause cell death or proliferation defects in the dorsal neural tube. (a) Whole mount view of an HH9 embryo bilaterally transfected with control MO (blue) on the left and the Lin28a MO (green) on the right. Transverse sections showing the immunostaining for phosho-H3 (S10) (b) and Caspase-3 (c) in these embryos. (d–e) Transverse sections of HH9 embryos bilaterally transfected with let-7 mimic immunostained for phospho-H3 (d) or Caspase-3 (e). Quantification of average number of phosho-H3 (f) and Caspase-3 (g) positive cells on the neural tube/section in embryos transfected with Lin28a MO. Quantification of average number of phospho-H3 (h) and Caspase-3 positive (i) cells on the neural tube/section in embryos transfected with let-7 mimic. (j–o) Loss of Lin28a during early development disrupts the formation of neural crest derived trigeminal ganglia. Lateral view of Dapi-stained HH15 embryo showing control (j) and Lin28a MO (m) sides (control images were flipped horizontally to facilitate comparison). Immunohistochemistry for neuronal marker Tuj1 in control (k) and morpholino treated (n) side of the embryo shows the morphology of the trigeminal ganglion (white arrows point to the base of the ganglia). Higher magnification images of the trigeminal ganglia on the control (l) and morpholino-transfected (o) side of the embryo show disorganization of the trigeminal ganglia (n = 6/8). The maxillomandibular branch (red arrows) is significantly thinner in the MO transfected side. The white arrows point to the neurons of the ophthalmic branch which are disorganised on the morphants. MO: Morpholino, ns: not significant.
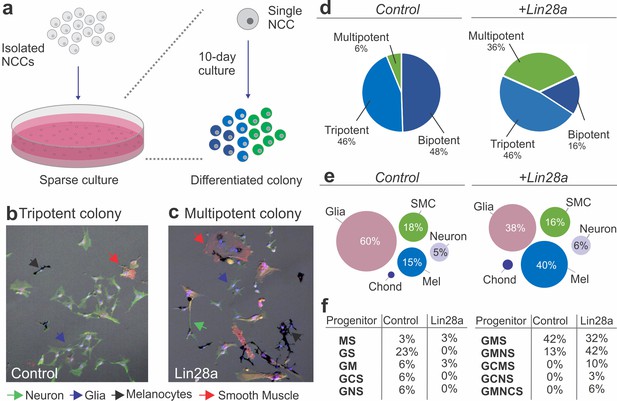
Lin28a modulates the developmental potential of neural crest cells.
(a) Diagram showing the experimental design for neural crest single-cell clonal analysis. Quail neural crest cells transfected with control or Lin28a expression construct were isolated and plated sparsely to ensure the formation of clonal colonies. Cells were cultured for up to 10 days, after which the different cell types in each colony were identified by immunofluorescence. (b–d) Neural crest cells expressing higher levels of Lin28a, formed increased number of multipotent colonies. Representative images showing a tripotent colony derived from a neural crest cell transfected with control construct (b), and a multipotent colony derived from a single neural crest cell transfected with a Lin28a expression construct (+Lin28a) (c). (d) Pie-charts showing the percentage of bi-, tri-, and multipotent colonies formed by control and +Lin28a neural crest cells. (e) Representation of the frequency of different cell types observed in all colonies formed by control and +Lin28a neural crest cells. (f) Table listing the percentages of the different neural crest progenitor cells observed in control and +Lin28 a conditions. NCC: neural crest cell, N: neuron, G: glia, M: melanocyte, C: chondroblast, S: smooth muscle cell.
-
Figure 2—source data 1
Quantitation of the different cell types in control vs +Lin28 neural crest derived colonies.
- https://doi.org/10.7554/eLife.40556.009
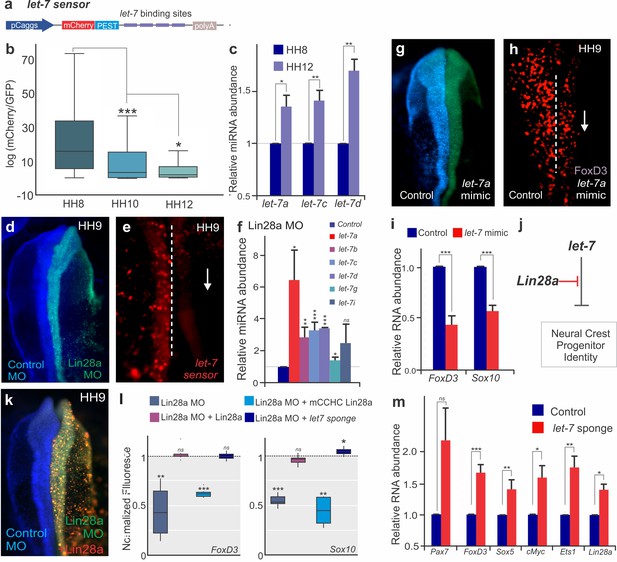
The Lin28/let-7 axis modulates neural crest progenitor identity in vivo.
(a) A schematic representation of the let-7 sensor, which consists of several let-7 binding sites downstream of destabilized mCherry fluorescent protein. (b–c) Activity of mature let-7 miRNAs increase through neural crest development. (b) Boxplots showing mCherry/GFP fluorescence ratio, a readout of let-7 sensor activity, in neural crest cells at different developmental stages. (c) RT-PCR for mature let-7 family miRNAs comparing their levels in neural crest cells sorted from HH8 and HH12 embryos. (d–f) Loss of Lin28a results in increased activity of mature let-7 miRNAs. (d) Whole mount view of an embryo bilaterally injected with control and Lin28a MO. (e) Representative image showing let-7 sensor fluorescence in control vs Lin28a MO side of an embryo. Dotted line represents embryo midline. (f) RT-PCR for mature let-7 family miRNAs, in the background of Lin28a knockdown. (g) Whole mount view of an embryo electroporated with control and let-7a mimic. (h) Immunohistochemistry for FoxD3 positive neural crest cells in the presence of let-7a mimic. Dotted line represents embryo midline. (i) Quantification of transcript levels of FoxD3 and Sox10, in presence of increased let-7a. (j) Model for modulation of neural crest identity by the Lin28/let-7 axis. (k) Representative dorsal view of an embryo electroporated with control MO (blue) on the left and Lin28a MO (green) co-injected with a Lin28a expression vector (red) on the right. (l) Boxplots showing the quantification of FoxD3 and Sox10 fluorescence in epistatic experiments, in which Lin28a Mo was co-electroporated with Lin28a expression vector, mCCHC Lin28a, and a let-7 sponge construct. (m) Loss of let-7 activity results in maintenance of multipotency genes in late neural crest cells. RT-PCR for Pax7, FoxD3, Sox5, Myc, Ets1 and Lin28a comparing the expression of these genes in control vs late migratory neural crest cells expressing let-7 sponge construct. Error bars in (c), (f), (i) and (m) represent standard error. HH: Hamburger and Hamilton developmental stages, MO: Morpholino.
-
Figure 3—source data 1
Data for the RT-PCR experiments shown in Figure 3, and quantitation of FoxD3 and Sox10 intensity in epistasis experiments.
- https://doi.org/10.7554/eLife.40556.011
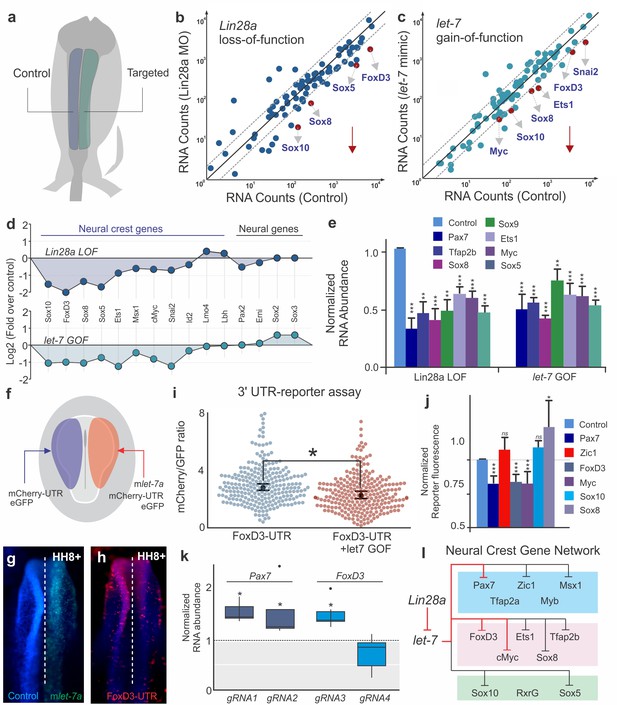
let-7 directly targets multipotency circuits in the neural crest transcriptional network.
(a) Control and targeted neural folds were dissected from the same embryo for Nanostring analysis. Comparison of transcript levels between (b) Control MO and Lin28a MO electroporated cells and (c) between control and let-7 mimic injected cells. Genes below the diagonal dotted line were significantly downregulated (downward red arrow) in each condition. (d) Neural crest genes were similarly affected in Lin28a loss of function (LOF) and let-7 gain-of function (GOF) assays. (e) RT-PCR performed with control and targeted neural folds to validate Nanostring results. (f) Electroporation scheme for in vivo 3’UTR reporter assay. Individual 3’UTR reporter constructs were co-injected with a control oligo (left) and a let-7 mimic (right). Flow cytometry analysis was performed to measure mCherry and eGFP fluorescence of individual cells. (g–h) whole mount view of an embryo showing FoxD3-UTR reporter fluorescence in control vs let-7 mimic transfected side of the embryo. (i) Representative scatter plots of FoxD3 UTR-reporter assay, showing the mCherry/GFP intensity ratio in cells analyzed from the control (gene-UTR) and let-7a mimic transfected (gene-UTR +let7 GOF) sides of the same embryo. Each dot in the plot represents a single cell. (j) Average fold change in the ratio of mCherry/GFP intensity for each 3’UTR analyzed. (k) Quantification of fold change in Pax7 and Foxd3 transcript levels in late migratory neural crest cells when the let-7 binding site on the 3’-UTR of these genes are targeted with specific gRNAs. gRNA1 and gRNA2 against Pax7 3’-UTR specifically targets the two let-7 binding sites, while gRNA3 and gRNA4 for FoxD3 3’-UTR targets a let-7 binding site and another control region on the UTR, respectively. (l) Lin28/let-7 targets in the early neural crest transcriptional network, showing genes that are directly (red inhibitory lines) or indirectly (black inhibitory lines) affected by let-7. Error bars in (e) and (j) represent standard error and standard deviation respectively. MO: Morpholino, LOF: loss-of-function, GOF: gain-of-function.
-
Figure 4—source data 1
Raw counts of the Nanostring experiment and the data for the RT-PCR experiments shown in Figure 4.
- https://doi.org/10.7554/eLife.40556.014
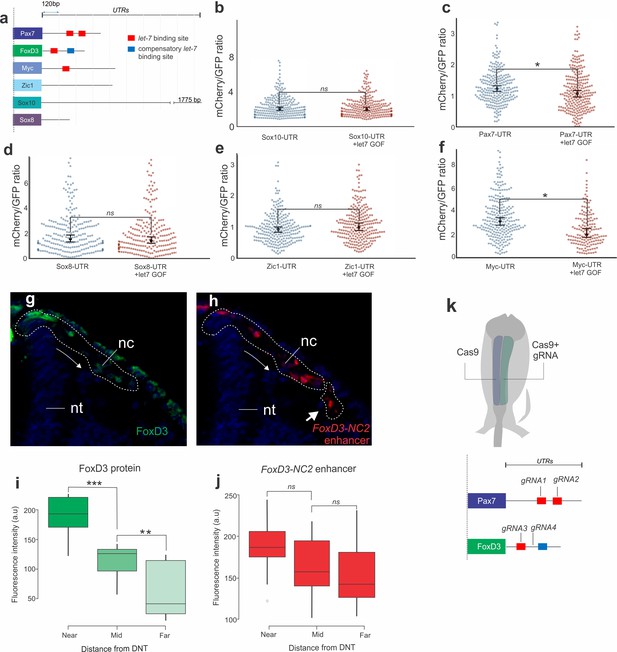
let-7 miRNAs regulate 3’-UTRs of neural crest genes.
(a) Position of let-7 binding sites in the 3’-UTRs of neural crest genes assessed in the reporter assay. Red boxes correspond to regions in the UTRs that are complementary to the let-7 seed sequence, while the blue boxes correspond to let-7 compensatory sites in the UTR, which are complementary to a region of the miRNA other than the seed sequence. Direct let-7 targets (Pax7, FoxD3, Myc) have seed sequence complementarity and, in the case of FoxD3, a compensatory binding site. These are absent in the UTRs of Sox10, Zic1, and Sox8. (b–f) Representative scatter plots of Sox10, Pax7, Sox8, Zic1 and cMyc UTR-reporter assay, showing the mCherry/GFP intensity ratio of each cell analyzed from the control (gene-UTR) and let-7a mimic transfected (gene-UTR +let7 GOF) halves of the same embryo. Each dot represents a single cell, and the medians and the 99% confidence intervals are overlayed on the scatter plots. (g–j) Single cell measurement of FoxD3 protein and NC2 mCherry-PEST reporter construct fluorescence in migrating NC cells. Transverse section of HH12 embryos, showing FoxD3 protein immunostaining (g) and FoxD3-NC2 enhancer reporter construct (h) in NC cells. Dotted lines show the migrating NC cells. The farthest migrated cells expressing FoxD3-NC2 enhancer (lower dotted region on (h)) are not positively stained for FoxD3 immunostaining, indicating decreased protein in these cells (g) Boxplots quantifying the fluorescent intensity of FoxD3 protein (i) NC2-mCherry-PEST reporter construct (j) in single neural crest cells as a function of the distance of the cells from the DNT. ‘NEAR’, ‘MID’ and ‘FAR’ corresponds to cells within 0–200 a.u, 201–350 a.u and 350–600 a.u from the DNT. (k) Bilateral electroporations of control vs. targeted Cas9 expression vectors were used to disrupt individual let-7 sites in the UTRs of the neural crest genes. UTR- Un-Translated Region, DNT- Dorsal Neural Tube, nt: neural tube, nc: neural crest, a.u- Arbitrary Units (as measured using ImageJ).
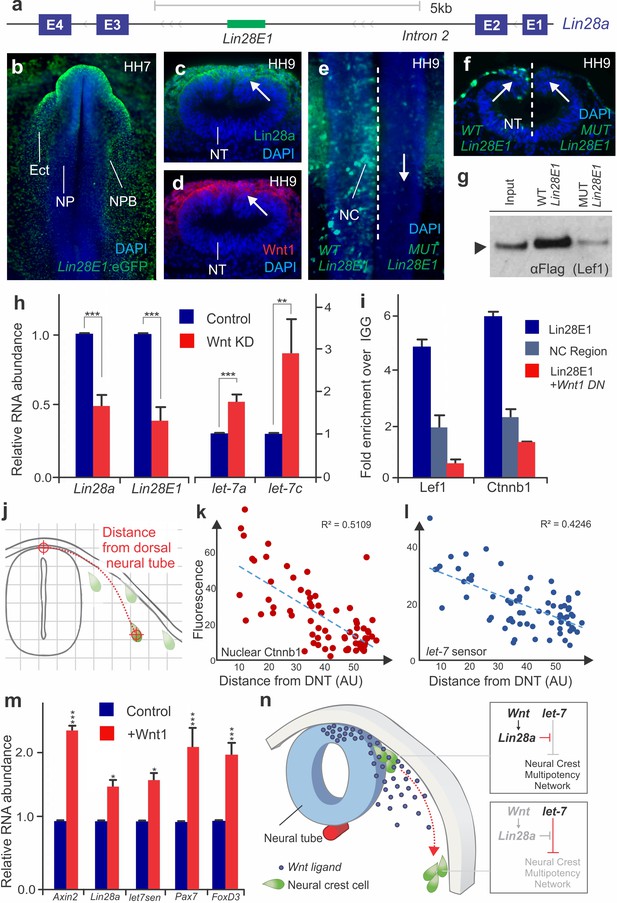
Positional information determines silencing of multipotency during neural crest migration (a) The avian Lin28a gene locus, showing the exonic (E1–E4) and intronic regions.
Lin28E1 is a 698 bp region within the second intron, which was identified as a Lin28a enhancer. (b) Expression pattern of a Lin28E1 driven eGFP construct, which is active in the ectoderm (Ect) and at the neural plate border (NPB). Immunohistochemistry for Lin28a (c) and Wnt1 (d) on transverse sections of HH9 embryo (arrows point to the dorsal tube). (e–f) Comparison of Lin28E1 (left) and MUT Lin28E1 (right) reporter activity in a bilaterally electroporated embryo. Dorsal view of the head of a whole mount embryo (e) and transverse section (f, arrows point to neural crest cells). (g) Western blot for flag-Lef1 following enhancer pull down experiment with wild type and mutant Lin28E1. (h) RT-PCR for endogenous Lin28a mRNA, Lin28E1 reporter and mature let-7 miRNAs on combinatorial knockdown of Wnt1 and Wnt4 (i) Chromatin immunoprecipitation for Lef-1 and β-catenin (Ctnnb1), performed with neural folds of WT embryos, and embryos electroporated with Wnt dominant negative construct. (j) Diagram outlining the parameters for single cell measurements of let-7 sensor fluorescence and nuclear β-catenin (Ctnnb1), as a function of the distance to the dorsal neural tube. Fluorescence intensity of nuclear β-catenin (k) and let-7 sensor (l) is inversely correlated with the distance of the neural crest cell from the neural tube. In both graphs, each dot represents a single cell. (m) Quantitative comparison of transcript levels of Axin2, Lin28a, let-7 sensor, Pax7 and FoxD3 in control vs. Wnt1 overexpressing neural crest cells (n) Model summarizing the results. AU: arbitrary units, DN: dominant negative, DNT: dorsal neural tube, IGG: Immunoglobulin G, Lin28E1: Lin28a Enhancer 1, Neg region: negative control region. Error bars in (h) and (m) represent standard error and error bars in (i) reflect standard deviation between technical replicates.
-
Figure 5—source data 1
Data for the RT-PCR experiements shown in Figure 5 and quantification of single cell measurements of let-7-sensor activity and nuclear b-catenin.
- https://doi.org/10.7554/eLife.40556.018
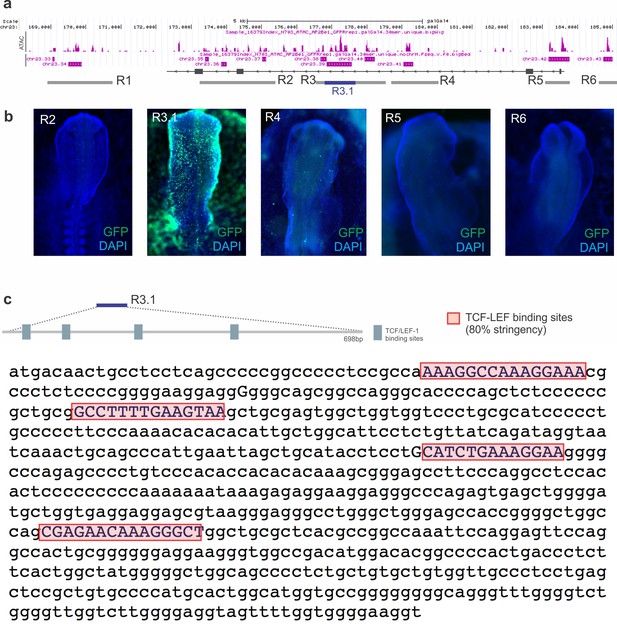
Identification of a Lin28a intronic enhancer.
(a) Genome browser snapshot of the ATAC seq peaks on chromosome 23 (Galgal 4.0) around and within the Lin28a gene locus. R1-R6 corresponds to the six regions (consisting of multiple ATAC peaks) which were cloned and assayed for enhancer activity (b). Representative images demonstrate that only R3/R3.1 were able to drive GFP expression in the embryo, in a pattern similar to that of Lin28a expression. Further analysis showed that R3.1, a 698 bp region located in the second intron of Lin28a, was sufficient to drive reporter expression. R3.1 (which was then renamed Lin28E1) has 4 TCF/LEF1 binding sites, as predicted by bioinformatics analysis. (c) Sequence of R3.1 (Lin28E1) showing the four TCF/LEF1 binding sites demarcated with red boxes.
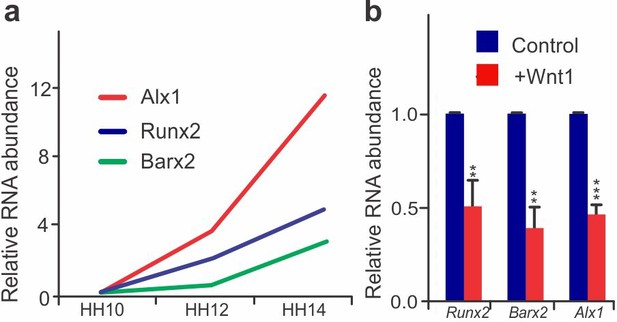
Prolonged Wnt signaling affects neural crest differentiation (a) Normalized expression levels of differentiation genes Alx1, Runx2, and Barx2 in migratory neural crest cells in different stages of development.
(b) RT-PCR comparing the levels of Runx2, Barx2 and Alx1 in control vs. Wnt1 overexpressing neural crest cells sorted from HH12 embryo. Error bars represent standard error.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse Anti-Lin28a | DSHB Cat # IE2 | RRID:AB_2618825 | IHC 1:4 |
| Antibody | Goat Anti-Sox10 | R and D Systems Cat# AF2864 | RRID:AB_442208 | IHC 1:50 |
| Antibody | Mouse Anti-Pax7 | DSHB | RRID:AB_528428 | IHC 1:4 |
| Antibody | Rabbit Anti-Cas9 | Takara | Cat #:632607 | IHC 1:200 |
| Antibody | Rabbit Anti-Wnt1 | Abcam Cat #: ab15251 | RRID:AB_301792 | IHC 1:200 |
| Antibody | Mouse Anti-Ctnnb1 | BD Biosciences Cat #:610154 | RRID:AB_397555 | IHC 1:100 |
| Antibody | Mouse Anti-Tuj1 | BioLegend, Cat #:801202 | RRID:AB_10063408 | IHC 1:200 |
| Antibody | Rabbit Anti-pH3(S10) | Abcam Cat# ab47297 | RRID:AB_880448 | IHC 1:200 |
| Antibody | Rabbit Anti Caspase-3 | R and D Systems Cat #: AF835 | RRID:AB_2243952 | IHC 1:100 |
| Antibody | Rabbit Anti-mCherry | Abcam Cat # ab167453 | RRID:AB_2571870 | IHC 1:200 |
| Recombinant DNA Reagent | pRNA-U6-let-7-sponge | Addgene | plasmid # 35664 | |
| Recombinant DNA Reagent | pCAGGS-let-7-mCherry-PEST sensor | this paper | Detailed in Materials and methods section | |
| Recombinant DNA Reagent | pX333 vector | Addgene | plasmid # 64073 | |
| Recombinant DNA Reagent | pCAGGS-Lin28a-H2B-RFP | ths paper | Detailed in Materials and methods section | |
| Recombinant DNA Reagent | pCAGGS-Lin28a-mCCHC-H2B-RFP | this paper | Detailed in Materials and methods section | |
| Recombinant DNA Reagent | pTK-Tfap2aE1-GFP | this paper | Detailed in Materials and methods section | |
| Recombinant DNA Reagent | pTK-Lin28E1-GFP | this paper | Detailed in Materials and methods section | |
| Sequence based reagent | Lin28a morpholino | Genetools (this paper) | 5’-AAACAGACCCCATCCCGACACTCGC-3’ | |
| Sequence based reagent | Wnt1 morpholino | Genetools (this paper) | 5’-GATGATGCCCCTACGGAGCGGGAAT-3’ | |
| Sequence based reagent | Wnt4 morpholino | Genetools (this paper) | 5’-GCGCAGGAAATACTCCGGGCTCATC-3’ | |
| Sequence based reagent | Lin28a gRNA1 | this paper | 5'-ACCGATACCCTCAAAGCTGGCCGAGG-3' | |
| Sequence based reagent | Lin28a gRNA2 | this paper | 5'-CACCGCGCTTGCAA ATTCCGAGTTGTGG-3' | |
| Sequence based reagent | LIn28a DsiRNA1 | IDT (this paper) | 5’GCCGUUGAAUUCACCUUCAAGAAAT-3’ | |
| Sequence based reagent | Lin28a DsiRNA2 | IDT (this paper) | 5’-GGGGUCUGUUUCCAACCAGCAGUTT-3’ | |
| Sequence based reagent | LIN28b DsiRNA1 | IDT (this paper) | 5’-GUGGAAUUUACUUACAAGAAAUCTT-3’ | |
| Sequence based reagent | Lin28B DsiRNA2 | IDT (this paper) | 5’-AAGCUUACAUGGAAGGAUUUAGAA-3’ | |
| Commercial assay or kit | qScript microRNA cDNA Synthesis Kit | Quanta Biosciences | Cat #: 95107–025 | |
| Commercial assay or kit | Rneasy Plus Micro kit | Qiagen | Cat #: 74034 | |
| Commercial assay or kit | Power SYBR Green Cells-to-CT Kit | Thermo Fisher | Cat #: A35379 | |
| Software, algorithm | ImageJ/Fiji | NIH | ||
| Software, algorithm | nSolver | Nanostring technologies |
Additional files
-
Supplementary file 1
Table listing the number of biological replicates analyzed the calculated p-values for the quantitative experiments performed in this paper.
The corresponding figure number for each experiment is also included in the table.
- https://doi.org/10.7554/eLife.40556.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40556.020





