Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis
Figures
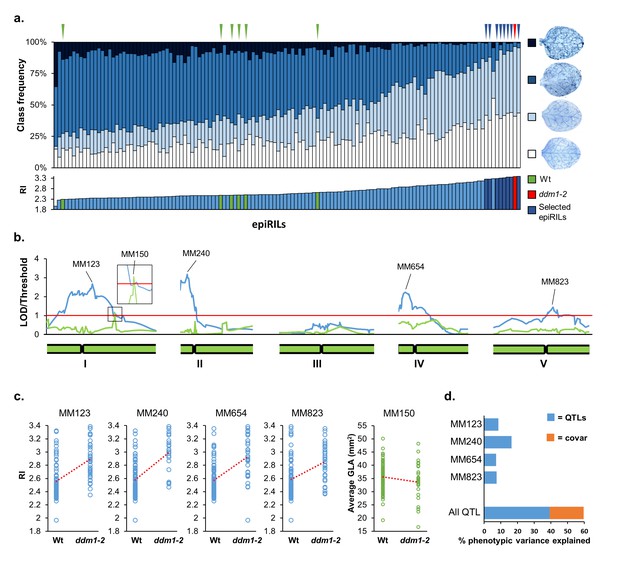
Mapping of epigenetic quantitative trait loci (epiQTL) controlling transgenerational resistance against Hyaloperonosopra arabidopsidis (Hpa).
(a) Levels of Hpa resistance in 123 epiRIL lines, the ddm1-2 line (F4; red triangle) and six Wt lines (Col-0; green triangles). Top graph shows distribution of infection classes in each epiRIL; blue triangles pinpoint the eight most resistant epiRILs with statistically similar levels of Hpa colonisation as the ddm1-2 line (Pearson’s Chi-squared test, p>0.05). Bottom graph shows variation in Hpa resistance index (RI). Green bars: Wt lines; red bar: ddm1-2; blue bars eight most resistant epiRILs (n > 100). (b) Linkage analysis of RI (blue line) and green leaf area (GLA) of three-week-old seedlings (green). Green bars at the bottom represent chromosomes. Red line represents the threshold of significance. Peak DMR markers with the highest LOD scores are shown on top. (c) Correlation plots between peak marker haplotype (methylated Wt versus hypomethylated ddm1-2) and RI (blue) or GLA (green). (d) Percentages of resistance variance explained by the peak DMR markers, including covariance between markers (orange).
-
Figure 1—source data 1
- https://doi.org/10.7554/eLife.40655.013

Representative examples of infection classes used for quantification of Hpa resistance.
Shown are trypan blue-stained Arabidopsis leaves at 6 days after spray-inoculation with Hpa. White (class I), absent or minimal colonisation; light blue (class II),≤50% leaf area colonised by the pathogen; dark blue (class III), ≤75% leaf area colonised by the pathogen, presence of conidiophores; black (class IV), >75% leaf area colonised by the pathogen, conidiophores and abundant sexual spores. Green arrows indicate colonisation by pathogen hyphae; blue arrows indicate hyphae surrounded by trailing necrosis, red arrows indicate conidiophores, black arrows indicate sexual oospores. Insets on the right show higher magnifications of colonisation markers. Scale bar = 50 μm.

Average green leaf area (GLA) of the 123 epiRILs (light green), the ddm1-2 line (F4; red) and six Wt lines (Col-0; dark green).
Shown are average GLA values of three-week-old plants (±SEM).
-
Figure 1—figure supplement 2—source data 1
- https://doi.org/10.7554/eLife.40655.006
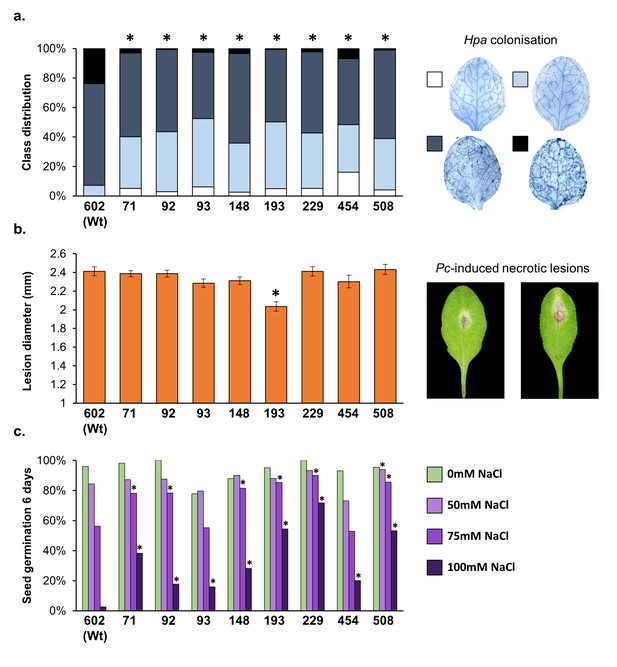
Resistance phenotypes of the eight most Hpa-resistant epiRILs against different (a) biotic stresses.
(a) Confirmation of resistance against biotrophic Hpa. Shown are levels of infection at 6 days post-inoculation (dpi) of 3-week-old plants. Trypan blue-stained leaves were analysed by microscopy and assigned to 4 Hpa infection classes (insets on the right; see Figure 1—figure supplement 1 for further details). Statistically significant differences in class distribution (asterisks) were analysed using Pearson’s Chi-squared tests (p<0.05) in pairwise comparisons with Wt line (#602); n > 80. (b) Quantification of resistance against necrotrophic Pletosphaerella cucumerina (Pc). Shown are average lesion diameters (±SEM) at 9 days after droplet inoculation with Pc spores onto similarly aged leaves of 5-week-old plants. Insets show representative examples of necrotic lesions by Pc. Statistically significant differences in necrotic lesions diameter (asterisks) were quantified by two-tailed Student’s t-test (p<0.05) in pairwise comparisons with Wt line (#602); n = 40–48. (c) Quantification of salt (NaCl) tolerance. Shown are percentages of seedlings developing full cotyledons after 6 days of growth on agar with increasing NaCl concentrations. Statistically significant differences in germination rates (asterisks) were quantified by Fisher’s exact test (p<0.05) in pairwise comparisons with Wt line (#602) at each salt concentration; n > 50.
-
Figure 1—figure supplement 3—source data 1
- https://doi.org/10.7554/eLife.40655.008

Defence marker phenotypes of the eight most Hpa-resistant lines.
(a) Relative expression of SA-dependent PR1 at 48 and 72 hpi with Hpa (red) or water (blue). Shown are mean relative expression values (±SEM). Statistically significant differences in relative expression (asterisks) were quantified by two-tailed Student’s t-test (p<0.05) in pairwise comparisons with Hpa-treated Wt line (#602); n = 3 (b) Resistance efficiency of callose deposition in Hpa-inoculated plants. Shown are percentages of arrested (light) and non-arrested (dark) germ tubes at 48 hpi. Insets show representative examples of aniline-blue/calcofluor-stained leaves by epi-fluorescence microscopy (bars = 100 μm; yellow indicates callose; blue indicates Hpa). Statistically significant differences in resistance efficiency of callose (asterisks) were analysed using Pearson’s Chi-squared tests (p<0.05) in pairwise comparisons with Wt line (#602); n > 150.
-
Figure 1—figure supplement 4—source data 1
- https://doi.org/10.7554/eLife.40655.010
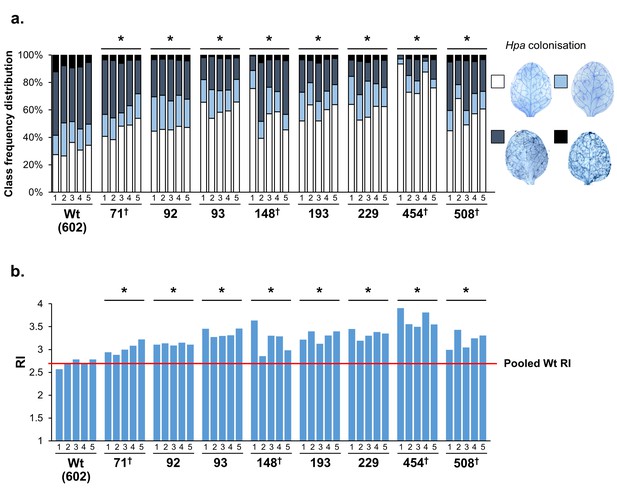
Transgenerational stability of Hpa resistance in Hpa-resistant epiRILs.
Five individual F9 plants from the eight most resistant epiRILs and the Wt line (#602) were self-pollinated to generate 40 F10 families. Plants of each F10 family were analysed for Hpa colonisation at 6 dpi. (a) Shown are frequency distributions of leaves across 4 Hpa colonisation classes (insets on the right; see Figure 1—figure supplement 1 for further details). (b) Resistance index (RI) values of the F10 families. The red line indicates the average RI value of the Wt (#602). Asterisks at the top of each graph indicate statistically significant differences in class distribution between pooled F10 families of the epiRIL relative to pooled F10 families of the Wt line (Pearson’s Chi-squared test; p<0.05). Crosses (†) at the bottom of each graph indicate statistically significant differences between F10 families within each epiRIL (Pearson’s Chi-squared test; p<0.05).
-
Figure 1—figure supplement 5—source data 1
- https://doi.org/10.7554/eLife.40655.012
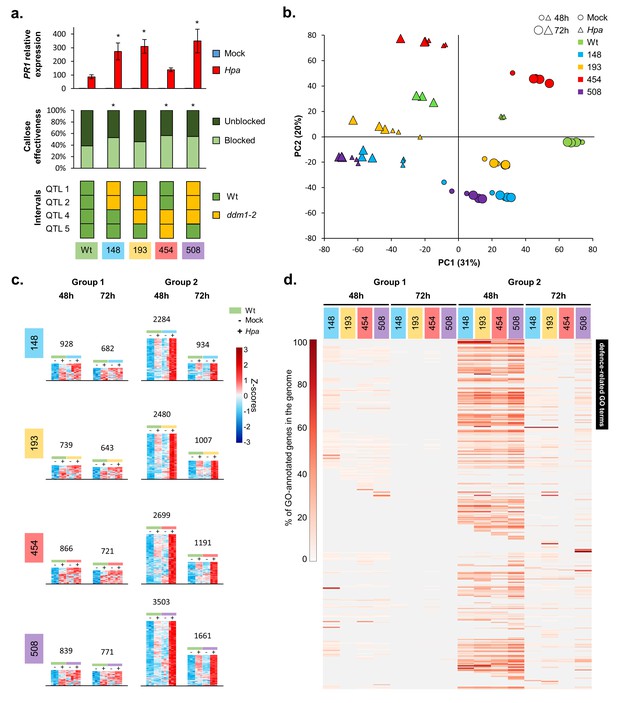
The defence-related transcriptome of Hpa-resistant epiRILs.
(a) Defence marker phenotypes and epiQTL haplotypes of 4 Hpa-resistant epiRILs and the Wt (#602), which were analysed by RNA sequencing. Top graph: relative expression of SA-dependent PR1 at 72 hr after inoculation (hpi) with Hpa (red) or water (blue). Middle graph: resistance efficiency of callose deposition in Hpa-inoculated plants. Shown are percentages of arrested (light) and non-arrested (dark) germ tubes at 48 hpi. Bottom panel: epiQTL haplotypes of selected lines. Green: methylated Wt haplotype; yellow: hypomethylated ddm1-2 haplotype. Asterisks indicate statistically significant differences to the Wt. (see Figure 1—figure supplement 4 for statistical information). (b) Principal component analysis of 27,641 genes at 48 (small symbols) and 72 (large symbols) hpi with Hpa (triangles) or water (Mock; circles). Colours indicate different lines. (c) Numbers and expression profiles of Hpa-inducible genes that show constitutively enhanced expression (Group 1) or augmented levels of Hpa-induced expression (Group 2) in the Hpa-resistant epiRILs at 48 or 72 hpi. Heatmaps show normalised standard deviations from the mean (z-scores) for each gene (rows), using rlog-transformed read counts (see Figure 2—figure supplements 3 and 4 for better detail) (d) GO term enrichment of primed and constitutively up-regulated genes. Shown are 469 GO terms (rows), for which one or more epiRIL(s) displayed a statistically significant enrichment in one or more categories (Hypergeometric test, followed by Benjamini-Hochberg FDR correction; q < 0.05). Heatmap-projected values for each GO term (rows) represent percentage of GO-annotated genes in each category relative to all GO-annotated genes in the Arabidopsis genome (TAIR10). Black bar on the top right indicates 111 defence-related GO terms.
-
Figure 2—source data 1
- https://doi.org/10.7554/eLife.40655.019
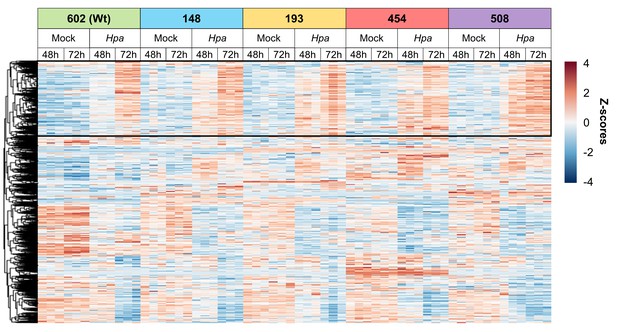
Hierarchical clustering of differentially expressed genes (DEGs) in selected Hpa-resistant epiRILs and the Wt at 48 and 72 hpi (Ward method).
The heatmap shows normalised standard deviations from the mean (z-scores) for each DEG (rows), using rlog-transformed read counts. Columns represent three biological replicates for each line-treatment-timepoint combination. The black square at the top of the heatmap indicates a cluster with genes that show augmented induction in one or more resistant epiRILs at 48 hr after Hpa inoculation.
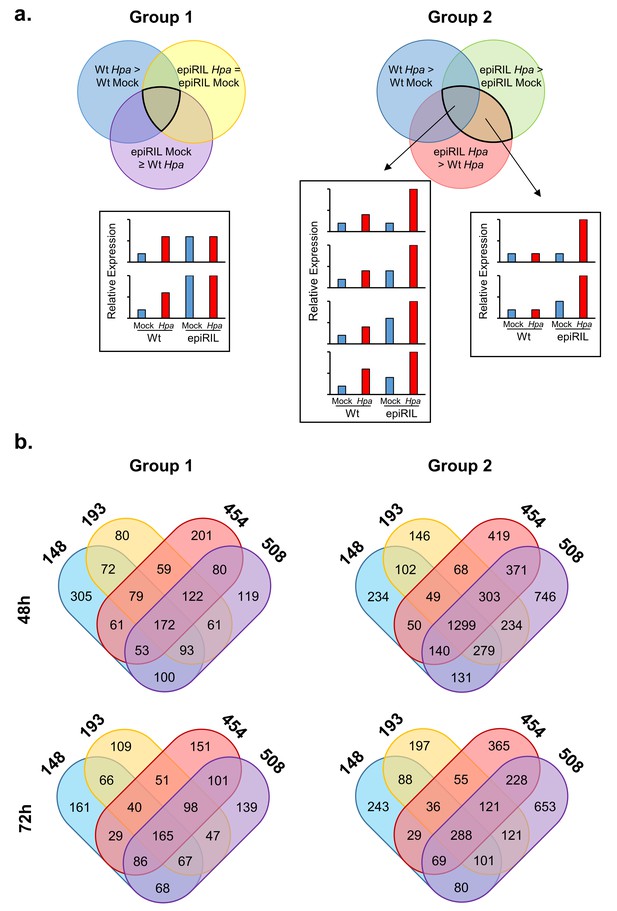
Selection of Hpa-inducible genes that show constitutively enhanced expression (Group 1) or enhanced Hpa-induced expression in the resistant epiRILs (Group 2).
(a) Description of each circle within the Venn diagrams indicates the statistical criteria used to obtain each selection (Wald test, q < 0.05). Overlapping areas in Venn diagrams (highlighted by black lines) indicate combinations of criteria used to select differently regulated genes. Panels below show schematic examples illustrating expression profiles of selected genes. (b) Number of Group 1 and Group 2 genes in Hpa-resistant epiRILs. Venn diagrams show numbers of genes that are unique or shared between epiRILs for each gene selection and time-point.
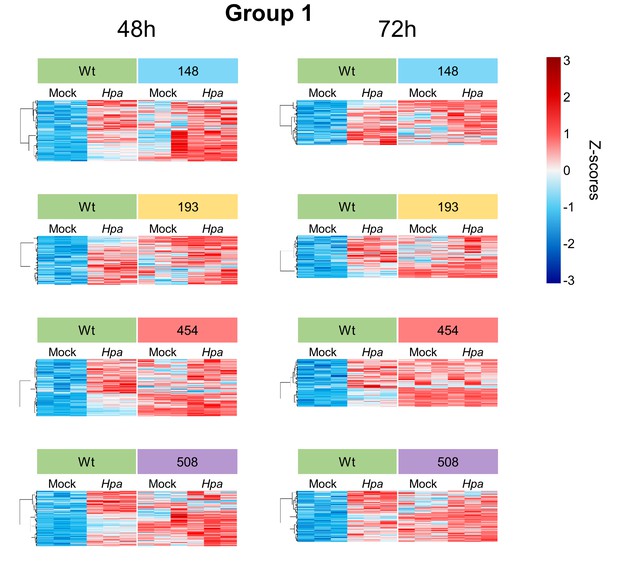
Transcript profiles of Hpa-inducible genes showing constitutively enhanced expression in the Hpa-resistant epiRILs (Group 1).
Heatmaps show normalised standard deviations from the mean (z-scores) for each gene (rows) at 48 and 72 hpi, using rlog-transformed read counts. Columns represent three biological replicates for each line-treatment combination. Expression profiles were subjected to hierarchical clustering by gene (Ward method).
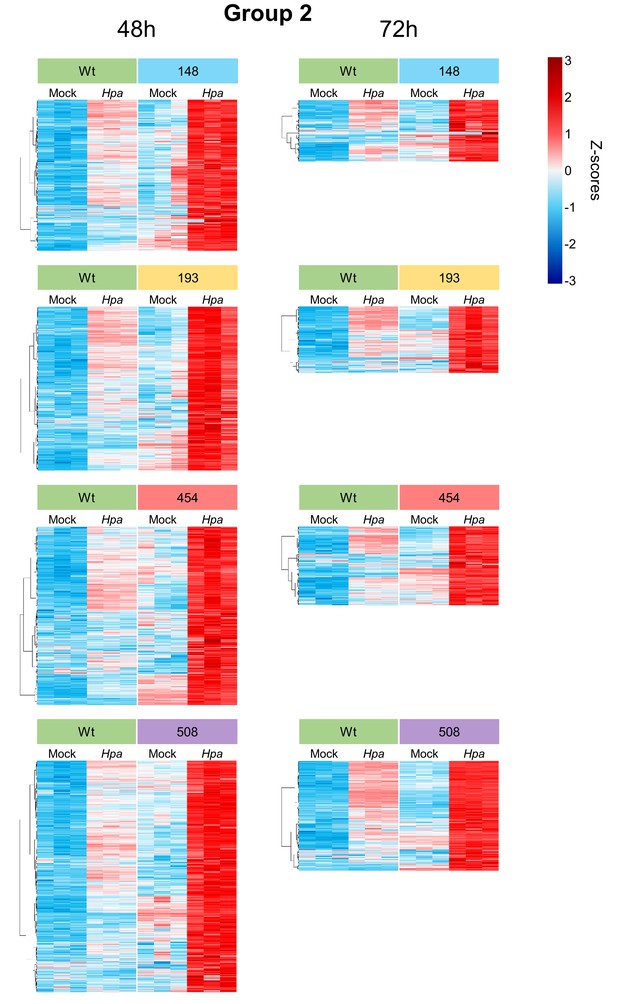
Transcript profiles of Hpa-inducible genes showing enhanced levels of Hpa-induced expression in the Hpa-resistant epiRILs (Group 2).
Heatmaps show normalised standard deviations from the mean (z-scores) for each gene (rows) at 48 and 72 hpi, using rlog-transformed read counts. Columns represent three biological replicates for each line-treatment combination. Expression profiles were subjected to hierarchical clustering by gene (Ward method).
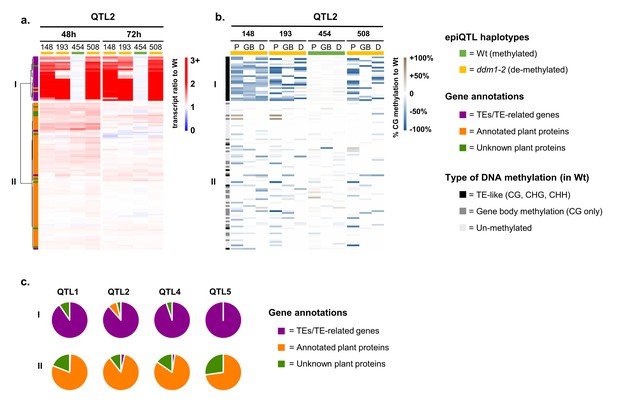
Relationship between augmentation of pathogen-induced expression and DNA methylation for epiQTL-localised genes.
(a) Expression profiles of epiQTL-based genes showing elevated levels of Hpa-induced expression in one or more epiRIL(s) (Group 2). Shown are genes located in the epiQTL interval of chromosome II (epiQTL2; LOD drop-off = 2; see Figure 3—figure supplement 1a for other the epiQTLs). Heatmap shows gene expression ratios between Hpa-inoculated epiRILs and the Wt, representing augmented expression levels during pathogen attack. Hierarchical clustering yielded two distinctly regulated gene clusters (I and II). Coloured bars on the top indicate epiQTL2 haplotypes. Green: methylated Wt haplotype. Yellow: hypomethylated ddm1-2 haplotype. (b) Levels of CG DNA methylation of the same genes in the epiQTL2 interval (see Figure 3—figure supplement 1b for other epiQTLs). Heatmap shows percentages of hypomethylation (blue) or hyper-methylation (brown) relative to the Wt for 2 kb promoter regions (P), gene bodies (GB) and 1 kb downstream regions (D). (c) Distribution of gene annotations of distinctly regulated gene clusters for each epiQTL.
-
Figure 3—source data 1
- https://doi.org/10.7554/eLife.40655.026

Relationship between augmentation of pathogen-induced expression and DNA methylation for epiQTL-localised genes.
(a) Expression profiles of epiQTL-based genes with elevated levels of Hpa-induced expression in one or more epiRILs (Group 2). Shown are genes located in the epiQTL intervals (LOD drop-off = 2) of chromosomes I (epiQTL1), chromosome IV (epiQTL4) and chromosome V (epiQTL5). (b) Levels of DNA methylation of the same genes in epiQTL1, epiQTL4 and epiQTL5. For details, see legend to Figure 3.
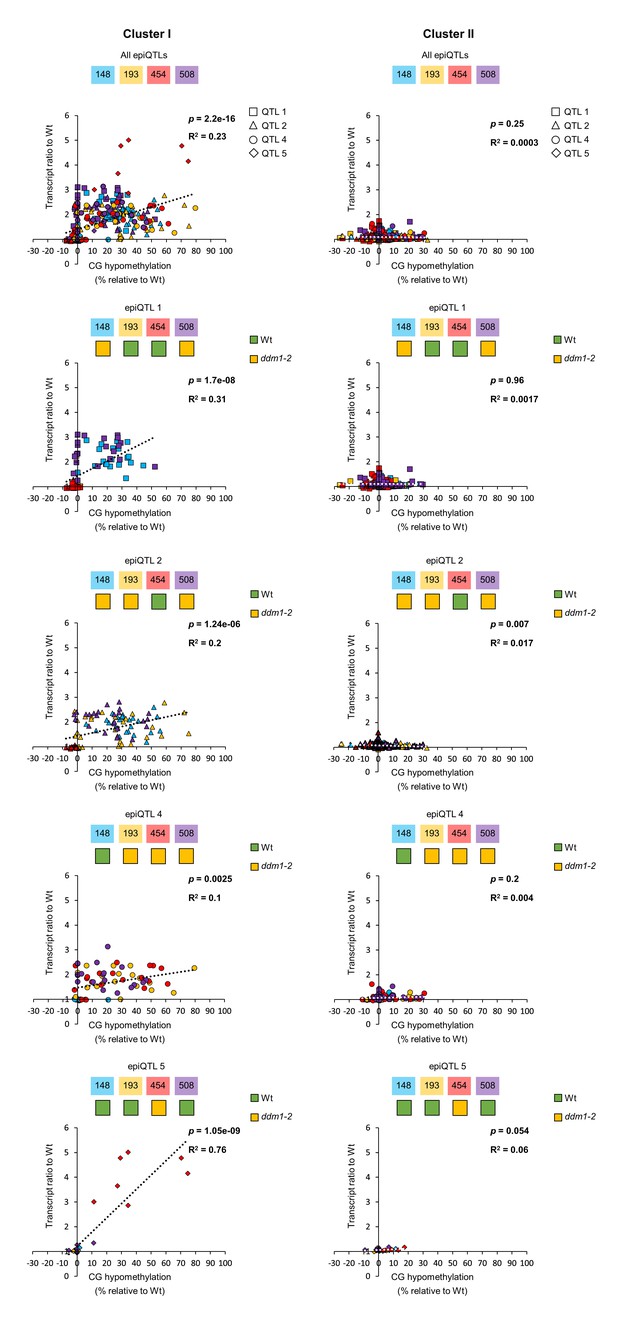
Correlation analysis between augmented gene transcription and DNA hypomethylation.
Augmented gene transcription was defined as the ratio between the Hpa-inoculated epiRIL and the Hpa-inoculated Wt at 48 hpi (Figure 3a and Figure 3—figure supplement 1a). DNA hypomethylation values were averaged across promoter regions, gene bodies and downstream regions. Scatter plots show transcript ratios against the hypomethylation for each gene in expression clusters I and II of Group 2 (Figure 3b and Figure 3—figure supplement 1b), which were selected by hierarchical clustering of augmented expression profiles in the epiRILs during Hpa infection. Significant positive correlations (Pearson linear regression; p<0.05) indicate cis-regulation by DNA methylation.
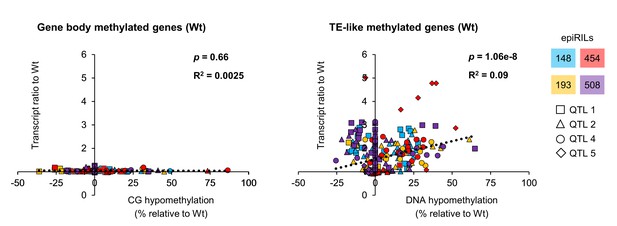
Correlation analysis between augmented gene transcription and type of DNA hypomethylation.
Scatter plots show augmented transcript ratios against hypomethylation for all epiQTL-based genes in Group 2 (Figure 3 and Figure 3—figure supplement 1). Augmented transcription was defined as the ratio between the Hpa-inoculated epiRIL and the Hpa-inoculated Wt at 48 hpi (Figure 3a and Figure 3—figure supplement 1a). Hypomethylation values at gene bodies in the epiRILs were divided according to the type DNA methylation. If hypomethylation occurred at CG context only, genes were classified as being reduced in gene body methylation (gbM); if hypomethylation occurred all three sequence contexts (CG, CHG, CHH), genes were classified as being reduced in TE methylation (teM). Values of gbM hypomethylation are expressed as percentage reduction in GC methylation relative to the Wt; values of teM hypomethylation are expressed as percentage reduction in all sequence contexts. Statistically significant correlations (Pearson linear regression; p<0.05) indicate cis-regulation by DNA methylation.

Genomic contexts of six plant protein-encoding genes in the epiQTL intervals, whose transcriptional priming coincides with reduced DNA methylation.
Orange bars indicate gene models; superimposed purple bars indicate associated transposable elements (TEs). Large blocks represent exons; lines between blocks represent introns; smaller blocks at the 3’ and 5’ ends represent un-translated regions. Units of the back scale correspond to 1 Kb.
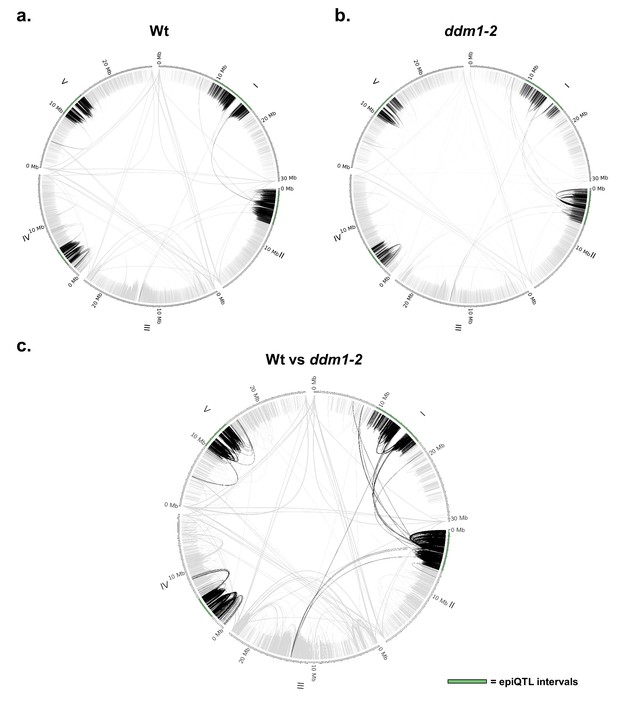
Genome-wide chromatin interactions in Wt and ddm1-2 Arabidopsis.
Circular diagrams show all five Arabidopsis chromosomes. The four epiQTL regions are highlighted in green. Chromatin interactions are indicated by lines. Gray lines: interactions outside the epiQTLs. Black lines: interactions with the epiQTLs. Presented results are based on Hi-C data from Feng et al. (2014) (Weber et al., 2016) (a) Genome-wide chromatin interactions in the Wt (Col-0). (b) Genome-wide chromatin interactions in the ddm1-2 mutant. (c) DDM1-dependent chromatin interactions that are altered in the ddm1-2 mutant compared to the Wt plants.
Additional files
-
Supplementary file 1
Supplementary datasets.
(S1) Distribution of 126 DMR markers for Wt and ddm1-2 haplotypes in all 123 epiRILs. (S2) Linkage between shared transposition events (STEs) in the epiQTL of Chromosome 1 and Hpa resistance. (S3) Differentially expressed genes between all epiRIL/treatment/time-point combinations. (S4) Genes showing statistically significant induction by Hpa in the Wt and/or Hpa-resistant epiRILs. (S5) Hpa-inducible genes displaying constitutively up-regulated expression in one or more Hpa-resistant epiRILs. (Group 1). (S6) Hpa-inducible genes displaying augmented induction in one or more Hpa-resistant epiRILs. (Group 2). (S7) Percentages of genes in Group 2 showing statistically signigicant interaction effect between epigenotype and treatment. (S8) Defence-related GO terms showing significant enrichment in Group 1 and/or Group 2 genes. (S9) Hpa-inducible genes whithin epiQTL intervals that do not show positive correlation between augmented induction and DNA hypo-methylation (trans-regulated). (S10) Hpa-inducible genes whithin epiQTL intervals that show positive correlation between augmented induction and DNA hypo-methylation (cis-regulated).
- https://doi.org/10.7554/eLife.40655.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40655.028





