Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility
Figures
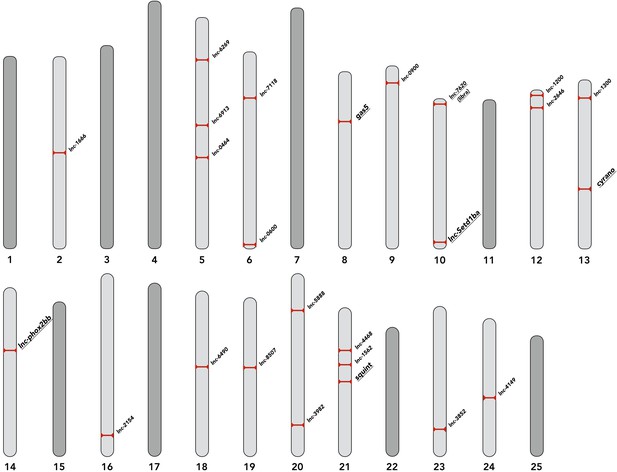
Genomic location of selected lncRNAs.
The chromosomal positions of selected lncRNAs are depicted. lncRNAs discussed in the text are underlined. The corresponding genomic coordinates for all lncRNAs are provided in the supplementary file 2.
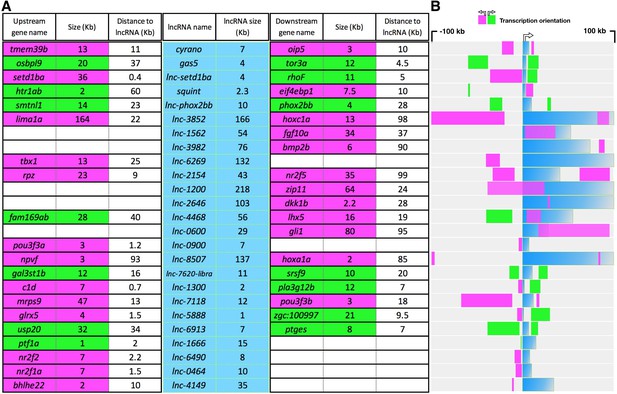
Size, relative distance and orientation of selected lncRNAs and their neighboring genes
(A) lncRNA names and sizes are shown in the middle section (blue columns). The distance, size and transcriptional orientation of the neighboring genes, in a 200 kb window centered on lncRNA’s TSS are shown on the left (upstream neighbor) and on the right (downstream neighbor). The transcription orientation is represented by green (in the same direction as lncRNA) and magenta (in the opposite direction of lncRNA). (B) Visual representation of data in A. All sizes and distances are in Kb.
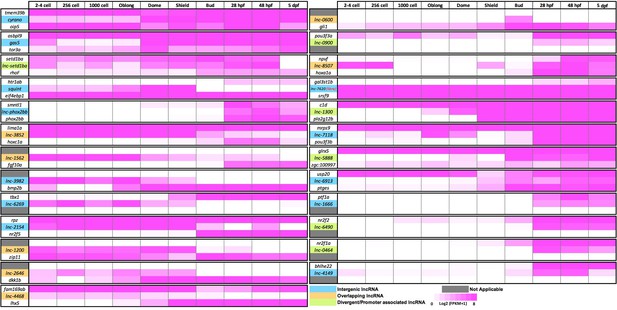
Expression levels of selected lncRNAs and their neighboring protein-coding genes.
LncRNAs are color coded as blue (Intergenic), brown (Overlapping) and green (Divergent/Promoter associated) (see Figure 1—figure supplement 1B). For each lncRNA and its upstream (top) and downstream (bottom) neighbor, the expression levels at 10 early-developmental stages are shown (Pauli et al., 2012). The scale is log2 (FPKM +1) value, represented as gradient between 0 (white) and 8 (magenta).
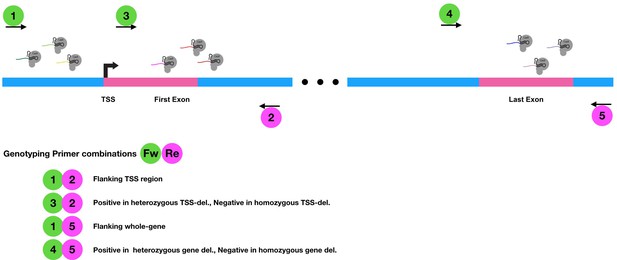
Cas9-mediated deletion approach for generating lncRNA knockouts 6 gRNAs (three at either side of the TSS) were used to remove TSS.
Nine guide RNAs (the first six plus three additional gRNAs around the Transcriptional Termination Site, TTS) were used to generate the gene deletions. Relative positions of genotyping primers are indicated by numbered circles.
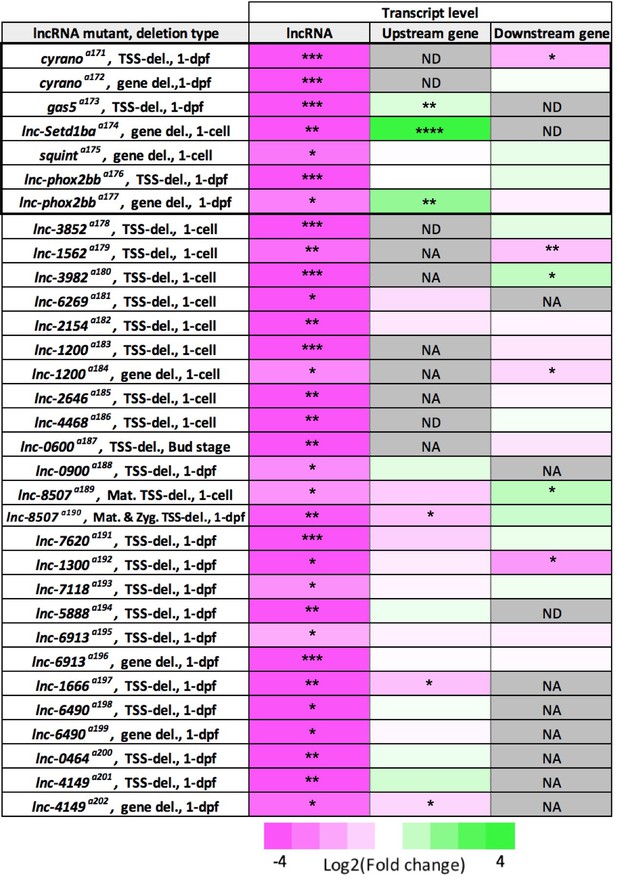
Summary of qRT-PCR analysis for lncRNA and their neighboring genes.
Visual representation of the expression level changes for each lncRNA and its neighboring genes in homozygous deletion mutants. Three biological replicates for homozygous mutant and wild-type samples. Log2 of fold change between −4 (magenta) and 4 (green) is shown.
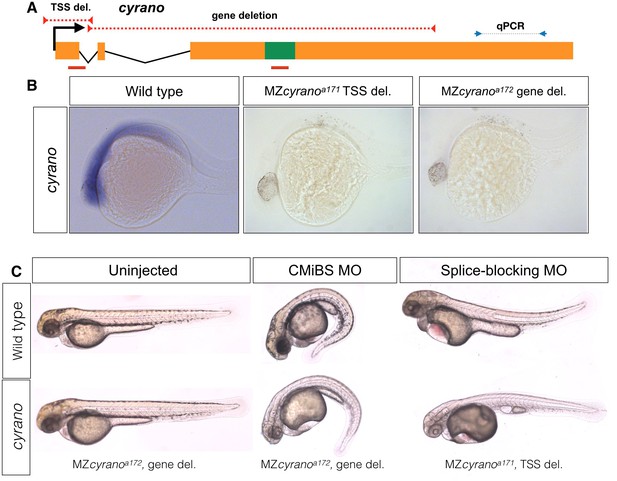
Normal embryogenesis of cyrano mutants.
(A) The positions of TSS-deletion allele and gene deletion allele are marked by dashed red lines. Green box represents the conserved element in cyrano which is complementary to miR-7. Solid red lines indicate the position of the first exon-intron boundary (e1i1) morpholino and conserved microRNA binding site (CMiBS) morpholinos. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) Representative images of in situ hybridization for cyrano in wild type (15/15) and both homozygous TSS-deletion (21/22) and gene deletion (18/18) 1-dpf. (C) At 2-dpf gene deletion mutants (lower-left), (and TSS-deletion mutants, not shown) were not different from the wild-type embryos (upper-left). Morpholino injected wild-type embryos (upper-middle and upper-left) reproduced observed phenotype in Ulitsky et. al (Kok et al., 2015). Morpholino injected deletion-mutants, lacking the corresponding binding sites for morpholinos, (lower-middle and lower-left) were comparable to morpholino injected wild types.
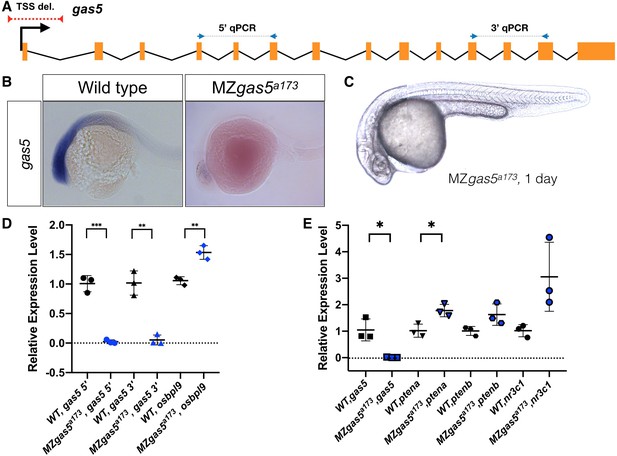
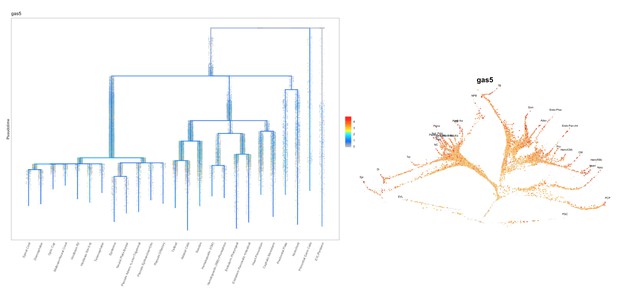
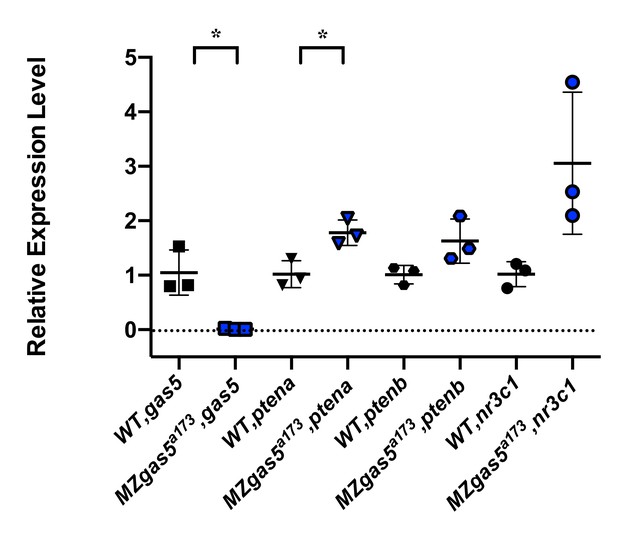
Normal embryogenesis of gas5 mutants.
(A) Position of the TSS-deletion allele in gas5 is marked by dashed red line. Arrows flanking black dotted lines mark the primer binding sites for 5’-qPCR and 3’-qPCR products. (B) Representative in situ hybridization images for gas5 in wild type (11/11) and homozygous TSS-deletion mutants (11/11). (C) Maternal and Zygotic gas5 (MZgas5) mutant embryos at 1-dpf were indistinguishable from the wild-type embryos at the same developmental stage (not shown). (D) Expression level of gas5 and osbpl9 measured by qRT-PCR. Tor3A, the other neighboring gene, was not expressed at the investigated time-point. (E) Expression level of gas5, its trans targets ptena, ptenb and nr3c1 measured by qRT-PCR. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (*, **, ***, and **** respectively mark p-values<0.05,<0.01,<0.001 and<0.0001).
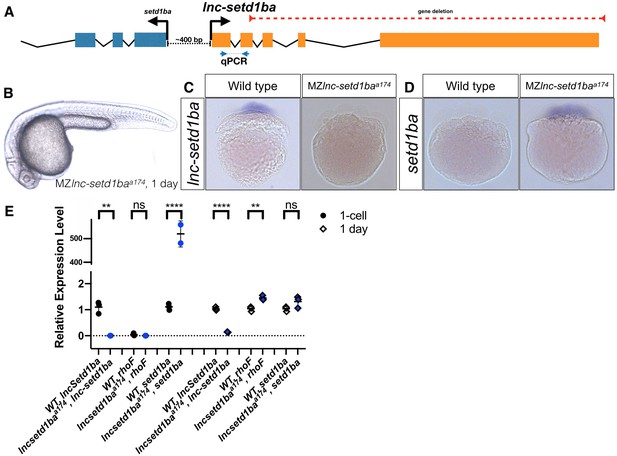
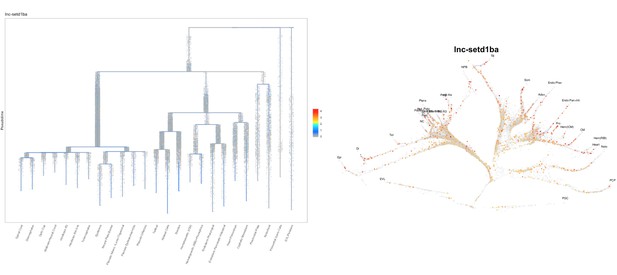
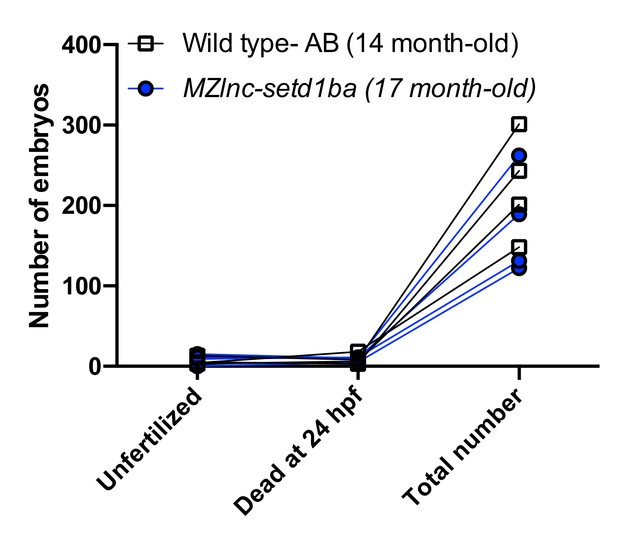
Normal embryogenesis of lnc-setd1ba mutants.
(A) The relative position of lnc-setd1ba and the protein-coding gene setd1ba. The gene deletion region is marked by dashed red line. Arrows flanking black dotted line mark the primer-binding sites for qRT-PCR product. (B) Maternal and zygotic lnc-setd1ba mutants were not different from wild-type embryos at 1-dpf. (C) Representative images of in situ hybridization for lnc-setd1ba at four- to eight-cell stage mutant (18/18) and wild-type (25/25) embryos. (D) In situ hybridization for the protein-coding mRNA, setd1ba (9/11) in lnc-setd1ba mutants compared to the wild-type embryos (15/15). (E) qRT-PCR at 1 cell stage and 1-dpf for the lncRNA and its neighboring genes rhoF and setd1ba. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (ns, *, **, ***, and **** respectively mark p-values≥0.05,<0.05,<0.01,<0.001 and<0.0001).
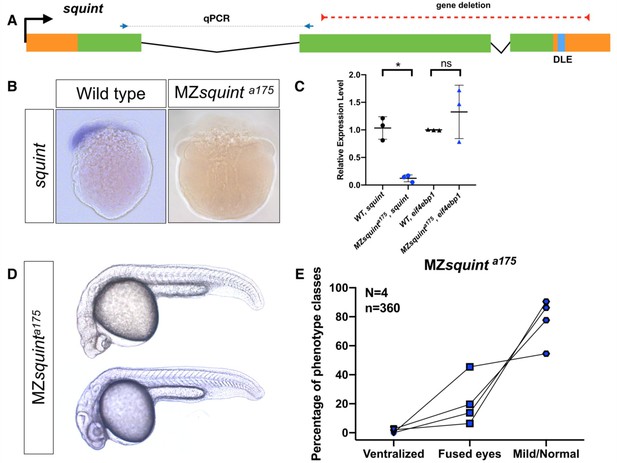
No non-coding function for squint 3’UTR.
(A) The position of untranslated regions (brown), coding region (green), putative Dorsal Localization Element- DLE (blue) and the gene deletion (red dashed line) in the squint genomic locus. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) In situ hybridization for squint at 8-cell stage on wild-type (18/20) and MZsquinta175(17/17) embryos. (C) qRT-PCR for squint and eif4ebp1 on wild-type and MZsquinta175 embryos at 1-cell stage. (D) Two representative MZsquinta175 embryos. (E) MZsquinta175 embryonic phenotype (N = 4 independent crosses, n = 360 embryos). The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (ns, *, **, ***, and **** respectively mark p-values≥0.05,<0.05,<0.01,<0.001 and<0.0001).
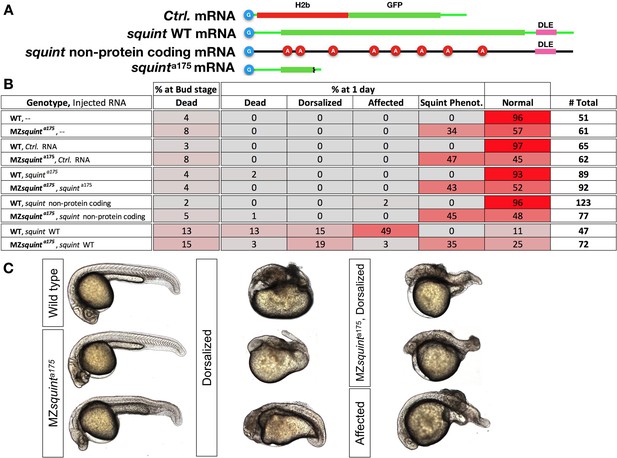
Dorsalization induced by Overexpression of squint mRNA but not its non-protein coding version.
(A) Schematic representation of injected mRNAs. Cap-analog is indicated by in blue circles at the beginning of each mRNA. squint non-protein coding mRNA was generated by adding 8 Adenine-nucleotides (red circles) after in-frame ATG codons. (B) Table shows scoring outcome of observed phenotypes in embryos injected with 30 pg of each indicated mRNA. (C) Representative embryos showing typical wild-type, squint mutant or dorsalized morphology. Ambiguous phenotypes were scored as ‘Affected’.
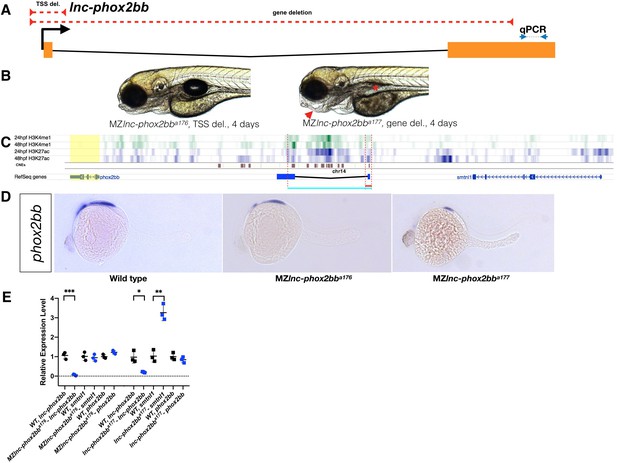
Requirement for lnc-phox2bb genomic elements but not RNA.
(A) The red dashed lines depict the respective positions of the lnc-phox2bb TSS and gene deletion. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) Homozygous gene deletion mutants but not the TSS-deletion mutants show embryonic defects in jaw formation (arrow head) and swim bladder inflation (asterisk) by 4-dpf. (C) Histone marks (H3K4me1 and H3K27ac) associated with enhancer activity (Bogdanovic et al., 2012) and conserved noncoding elements (CNEs) (Hiller et al., 2013) overlap with gene deletion. (D) phox2bb expression pattern in the TSS and gene deletions. (E) qRT-PCR analysis on MZ TSS-deletion and gene deletion mutants. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (*, **, ***, and **** respectively mark p-values<0.05,<0.01,<0.001 and<0.0001).
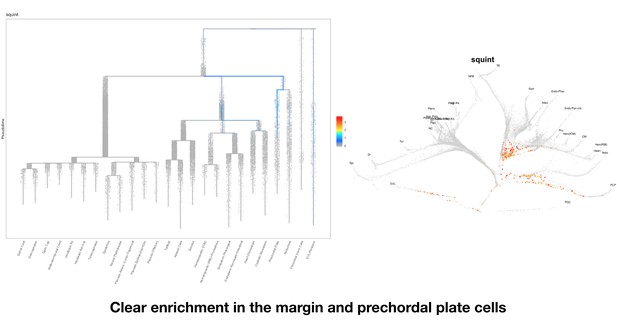
Clear enrichment in the margin and prechordal plate cells.
https://doi.org/10.7554/eLife.40815.021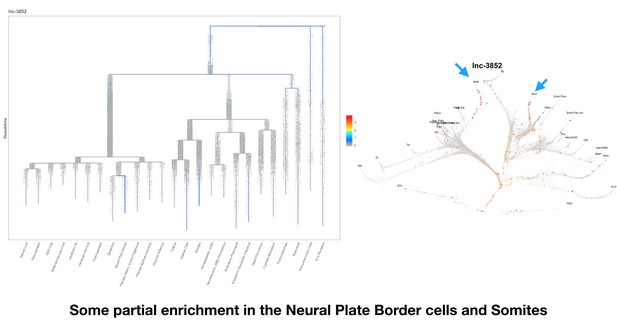
Some partial enrichment in the Neural Plate Border cells and Somites.
https://doi.org/10.7554/eLife.40815.022
Tables
Summary of lncRNA features and mutant phenotypes lncRNA names are shown in the first column.
lncRNAs were named using the last four digits of their corresponding ENSEMBL Transcript ID or their chromosome number if no transcript ID was available (e.g. lnc-1200 is located on chromosome 12). The second column represents ribosomal occupancy pattern along the length of lncRNAs in comparison to the 5’UTR, coding and 3’UTR of typical protein-coding transcripts (Chew et al., 2013). The third column shows the transcript ID for the investigated lncRNA or its genomic coordinate in GRCz10. Column Four shows the deletion size. Fifth column represent the percentage decrease in the level of lncRNA in comparison to wild type from three biological replicates (qRT-PCR). The six and seven columns show the presence of embryonic phenotypes, viability and fertility (at least 15 adult pairs per allele) of homozygous mutant fish. Eighth and ninth column show the upstream and downstream neighboring genes in a 200 kb window centered around the lncRNA’s TSS. The last column provides the selection criteria for each lncRNA.
| lncRNA mutant, deletion type | Ribosome Profiling, class | lncRNA transcript ID | Deletion size | Percent reduction | Embryonic phenotype | Viability and fertility | Neighboring genes | Selection criteria | |
|---|---|---|---|---|---|---|---|---|---|
| Up 100 Kb | Down 100 Kb | ||||||||
| cyranoa171, TSS-del. | Trailerlike | ENSDART00000139872 | 326 bp | 98% | No | Yes | tmem39b | oip5 | Syntenic and sequence conservation, Reported phenotype |
| cyranoa172, gene del. | Trailerlike | ENSDART00000139872 | 4374 bp | 94% | No | Yes | tmem39b | oip5 | Syntenic and sequence conservation, Reported phenotype |
| gas5a173, TSS-del. | Leaderlike | ENSDART00000156268 | 296 bp | 100% | No | Yes | osbpl9 | tor3a | Syntenic conservation, well studied lncRNA, host of several snoRNA |
| lnc-setd1baa174, gene del. | Leaderlike | ENSDART00000141500 | 3137 bp | 100% | No | Yes | setd1ba | rhoF | Syntenic and sequence conservation, Proximity to developmental regulatory genes |
| squinta175, gene del. | Coding | ENSDART0 0000079692 | 1032 bp | 95% | No | Yes | htr1ab | eif4ebp1 | Evolutionary conservation, Reported phenotype, putative cncRNA |
| lnc-phox2bba176, TSS-del. | Leaderlike | ENSDART00000158002 | 652 bp | 99% | No | Yes | smntl1 | phox2bb | Syntenic conservation |
| lnc-phox2bba177, gene del. | Leaderlike | ENSDART00000158002 | 9361 bp | 87% | Yes | No | smntl1 | phox2bb | Syntenic conservation |
| lnc-3852a178, TSS-del. | Leaderlike | ENSDART00000153852 | 447 bp | 100% | No | Yes | lima1a | hoxc1a | Maternal expression, Proximity to developmental regulatory genes |
| lnc-1562a179, TSS-del. | Leaderlike | ENSDART00000131562 | 409 bp | 90% | No | Yes | * | fgf10a | Maternal expression, Proximity to developmental regulatory genes |
| lnc-3982a180, TSS-del. | Leaderlike | ENSDART00000153982 | 352 bp | 97% | No | Yes | * | bmp2b | Maternal expression, Proximity to developmental regulatory genes |
| lnc-6269a181, TSS-del. | Leaderlike | ENSDART00000156269 | 535 bp | 99% | No | Yes | tbx1 | * | Maternal expression, Proximity to developmental regulatory genes |
| lnc-2154a182, TSS-del. | Trailerlike | ENSDART00000132154 | 546 bp | 100% | No | Yes | rpz | nr2f5 | Maternal expression, Proximity to developmental regulatory genes |
| lnc-1200a183, TSS-del. | Leaderlike | Chr12:1708389-1925779:1 | 590 bp | 95% | No | Yes | * | zip11 | Maternal expression, Longest selected lncRNA |
| lnc-1200a184, gene del. | Leaderlike | Chr12:1708389-1925779:1 | 203.8 kb | 84% | No | Yes | * | zip11 | Maternal expression, Longest selected lncRNA |
| lnc-2646a185, TSS-del. | Leaderlike | ENSDART00 000152646 | 240 bp | 97% | No | Yes | * | dkk1b | Proximity to developmental regulatory genes |
| lnc-4468a186, TSS-del. | Leaderlike | ENSDART00000154468 | 306 bp | 100% | No | Yes | fam169ab | lhx5 | Proximity to developmental regulatory genes, Low expression level |
| lnc-0600a187, TSS-del. | Trailerlike | Chr6:59414652-59443141:1 | 244 bp | 95% | No | Yes | * | gli1 | Proximity to developmental regulatory genes, Low expression level |
| lnc-0900a188, TSS-del. | Leaderlike | Chr9:6684669-6691350:1 | 377 bp | 83% | No | Yes | pou3f3a | * | Syntenic conservation, Low expression level |
| lnc-8507a189, mTSS-del. | Leaderlike | ENSDART00000158507 | 323 bp | 81% | No | Yes | npvf | hoxa1a | Proximity to Hox genes, Maternal and Zygotic promoters |
| lnc-8507a190, mzTSS-del. | Leaderlike | ENSDART00000158507 | 9773 bp | 95% | No | Yes | npvf | hoxa1a | Proximity to Hox genes, Maternal and Zygotic promoters |
| lnc-7620a191, TSS-del. | Trailerlike | ENSDART00000137620 | 668 bp | 99% | No | Yes | gal3st1b | srsf9 | Syntenic and sequence conservation, Implicated in adult fish and mouse behavior. Bitetti, A., et al. (2018) |
| lnc-1300a192, TSS-del. | Leaderlike | Chr13:4535992-4538275:1 | 367 bp | 92% | No | Yes | c1d | pla2g12b | Syntenic and sequence conservation, High expression level |
| lnc-7118a193, TSS-del. | Trailerlike | ENSDART00000157118 | 438 bp | 82% | No | Yes | mrps9 | pou3f3b | Syntenic conservation |
| lnc-5888a194, TSS-del. | Leaderlike | ENSDART00000155888 | 606 bp | 96% | No | Yes | glrx5 | zgc:100997 | Syntenic conservation, scaRNA13 host gene, shortest selected lncRNA |
| lnc-6913a195, TSS-del. | Leaderlike | ENSDART00000156913 | 333 bp | 72% | No | Yes | usp20 | ptges | Proximity to developmental regulatory genes |
| lnc-6913a196, gene del. | Leaderlike | ENSDART00000156913 | 5568 bp | 93% | No | Yes | usp20 | ptges | Proximity to developmental regulatory genes |
| lnc-1666a197, TSS-del. | Leaderlike | ENSDART00000141666 | 544 bp | 96% | No | Yes | ptf1a | * | Proximity to developmental regulatory genes, Restricted late expression |
| lnc-6490a198, TSS-del. | Leaderlike | ENSDART00000146490 | 607 bp | 99% | No | Yes | nr2f2 | * | Syntenic conservation, Restricted late expression |
| lnc-6490a199, gene del. | Leaderlike | ENSDART00000146490 | 8378 bp | 100% | No | Yes | nr2f2 | * | Syntenic conservation, Restricted late expression |
| lnc-0464a200, TSS-del. | Trailerlike | ENSDART00000140464 | 597 bp | 96% | No | Yes | nr2f1a | * | Restricted late expression pattern |
| lnc-4149a201, TSS-del. | Leaderlike | ENSDART00000154149 | 491 bp | 98% | No | Yes | bhlhe22 | * | Proximity to developmental regulatory genes |
| lnc-4149a202, gene del. | Leaderlike | ENSDART00000154149 | 35.11 kb | 100% | No | Yes | bhlhe22 | * | Proximity to developmental regulatory genes |
Additional files
-
Supplementary file 1
This compressed folder contains three Excel files for the sequences of gRNAs, genotyping and qRT-PCR primers (for lncRNAs and their neighboring genes) and also the annotated sequence files (.ape) for each lncRNA and their deleted segments.
- https://doi.org/10.7554/eLife.40815.014
-
Supplementary file 2
This genome-browser-compatible file is in the bed formant, containing the coordinates for all the lncRNAs investigated in this manuscript based on the GRCz11 (GCA_000002035.4).
- https://doi.org/10.7554/eLife.40815.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40815.016





