Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements
Figures
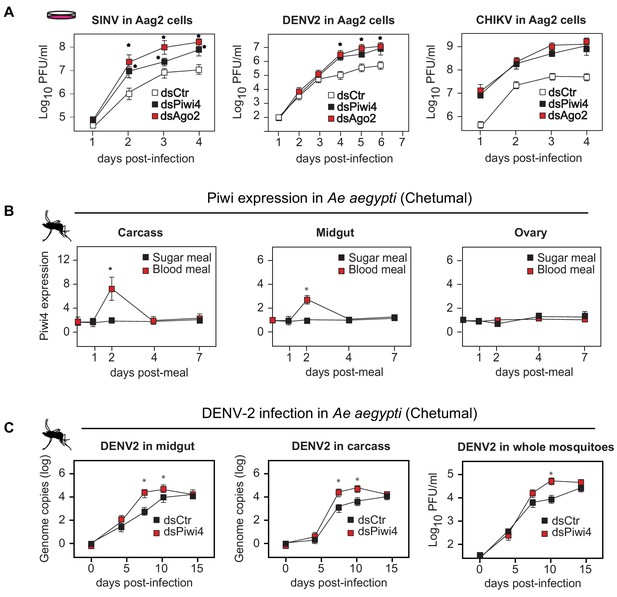
Piwi4 is a restriction factor in vivo, upregulated after blood meal.
(A) Multi step growth curves of SINV, DENV-2 and CHIKV in Aag2 cells (MOI 0.1) treated with dsRNA targeting Ago2 or Piwi4 measured by plaque assay. Error bars depict standard deviation of four replicates. Significant changes over controls are marked with asterisks (p<0.05, Mann-Whitney U test). (B) Piwi4 mRNA measured by RT-qPCR from pools of either blood- or sugar- fed Ae. aegypti mosquitoes. The mean and standard deviation of four biological replicates of pools of five mosquitoes in carcass, midgut and ovary are shown. Significant changes over controls are marked with asterisks (p<0.05, Mann-Whitney U test). (C) Replication of DENV2 as measured by RT-qPCR or virus plaque assays. Mosquitoes were infected by intrathoracic injection with either dsPiwi4 or control dsRNA prior to infection. Error bars correspond to standard error of 20 (RT-qPCR) or 30 (plaque assays) biological replicates of individual mosquitoes. Significant changes over controls are marked with asterisks (p<0.05, Mann-Whitney U test).
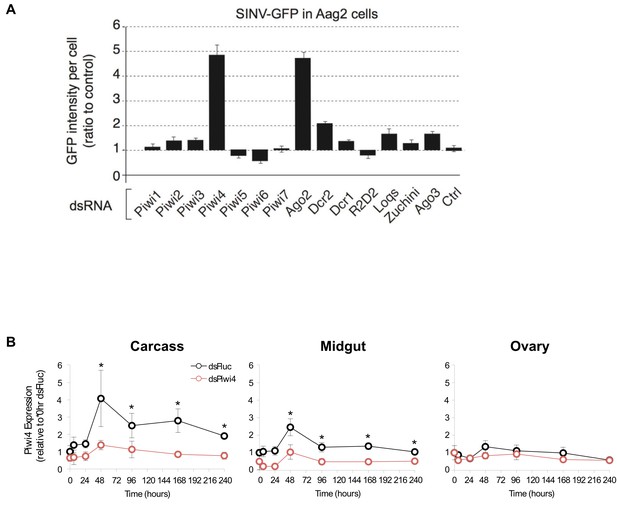
Piwi4 knockdown in cell culture and adult female Ae. aegypti mosquitoes.
(A) RNAi screen against the seven Aedes aegypti Piwi genes and other selected RNAi factors in SINV-eGFP infected Aag2 cells. Viral replication was measured as GFP intensity per cell. (B) Piwi4 mRNA measured by qPCR from pools of dissected tissues from blood-fed female Ae. aegypti mosquitos injected with dsRNA against Firefly luciferase (dsFluc, negative control) or against Piwi4 (dsPiwi4). The error bars represent standard deviations of four biological replicates of pools of five of the respective tissue. Significant changes over control are marked with asterisks (p≤0.05, Mann-Whitney U test).
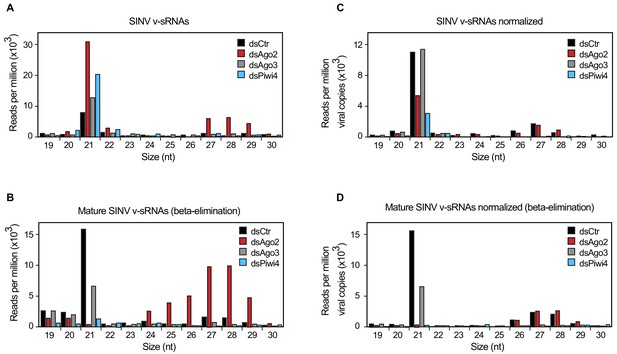
Piwi4 is required for v-piRNA maturation.
(A) Size distribution plot of small RNAs mapping to the SINV genome from infected Aag2 cells treated with Fluc/control, Piwi4, or Ago3 dsRNA. (B) Size distribution plot of beta-elimination resistant small RNA mapping to the SINV genome from infected Aag2 cells treated with Fluc/control, Piwi4, or Ago3 dsRNA. (C) Same as in (A) but read counts were normalized to viral copy numbers. (D) Same as in (B) but read counts for beta-elimination resistant small RNAs were normalized to viral copy numbers.
-
Figure 2—source data 1
Identification of small RNAs in the Aedes aegypti cell line Aag2 knocked-down for Ago2, Ago2 or Piwi4 expression and infected with Sindbis virus.
Aag2 cells were transfected with dsRNA against Firefly luciferase (control), Ago2, Ago3 or Piwi4, infected with Sindbis virus 2 days later and collected 3 days after infection for small RNA extraction.
- https://doi.org/10.7554/eLife.41244.007
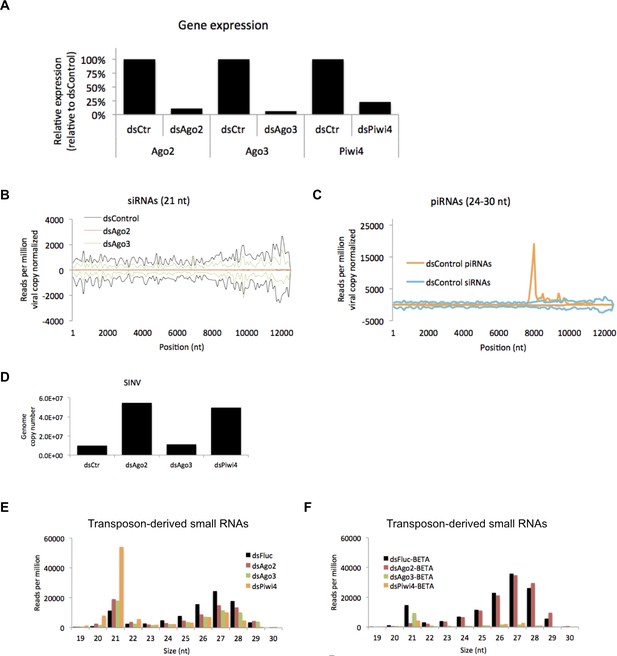
Effects of Ago2, Ago3 and Piwi4 knockdown on SINV-derived small RNAs.
(A) Ago2, Ago3, and Piwi4 mRNA measured by RT-qPCR from large RNA fraction of sample used for small RNA library preparation. (B) Mapping of siRNAs (21 nt) to SINV genome from SINV infected Aag2 cells treated with control (dsFluc), Ago2, Ago3 or Piwi4[O1] dsRNA. (C) Mapping of piRNAs (24-30 nt) to SINV genome from SINV infected Aag2 cells treated with control (dsFluc), Ago2, Ago3 or Piwi4 dsRNA. The number of small RNAs is normalized to the number of SINV genome copies for each dsRNA treatment. (D) SINV genomic RNA measured by RT-qPCR from large RNA fraction of sample used for small RNA library preparation. (E) Size distribution plot of small RNA mapping to transposons from infected Aag2 cell treated with control, Ago2, Ago3, or Piwi4 dsRNA. (F) Size distribution plot of beta-elimination resistant small RNA mapping to transposon from infected Aag2 cells treated with control, Piwi4, or Ago3 dsRNA.
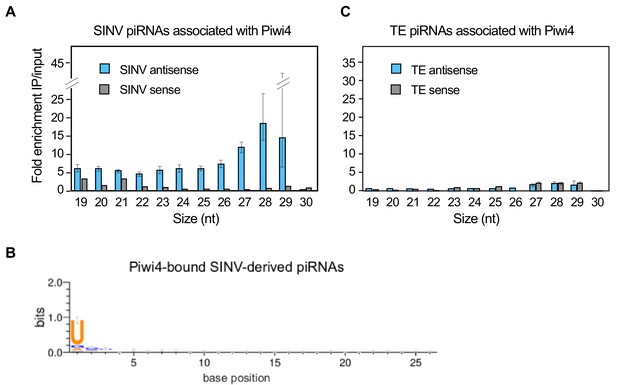
Piwi4 binds specifically to antisense v-piRNAs.
(A) Enrichment of SINV-derived small RNA in Piwi4-immunoprecipitation (IP) fraction compared to input sample. (B) The base bias for each position of SINV-derived piRNAs co-immunoprecipitated with Piwi4-FLAG shows a Uracil bias at position 1, characteristic of antisense Piwi-associated piRNAs (shown in bits). (C) Same as in A but for transposons (TE)-derived small RNAs.
-
Figure 3—source data 1
Identification of small RNAs preferentially bound by Piwi4 following Sindbis virus infection in the Aedes aegypti cell line Aag2.
Aag2 cells were transfected with plasmids expressing eGFP or FLAG-tagged Piwi4. Twenty-four hours after transfection, cells were infected with Sindbis virus and collected 3 days after infection and processed for IP of FLAG-Piwi4.
- https://doi.org/10.7554/eLife.41244.010
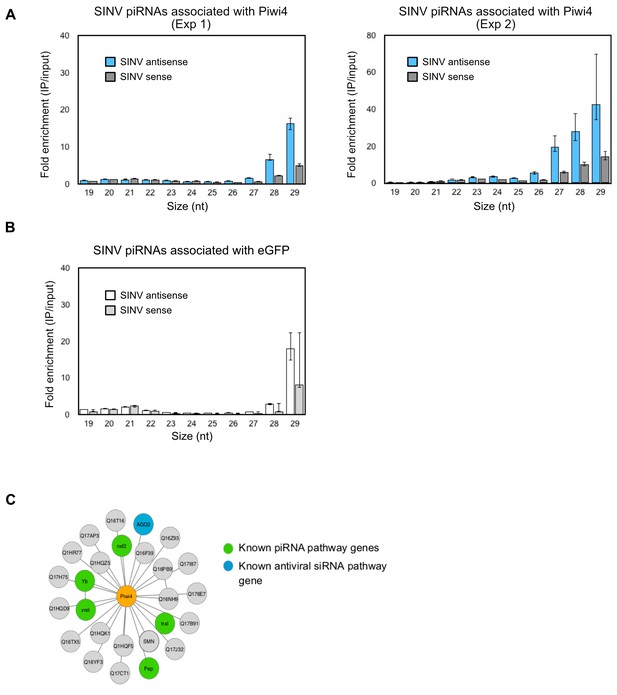
Piwi4 binds to bona-fide v-piRNAs and interacts with known siRNA and piRNA pathway components.
(A) Results from two independent biological replicates (Exp1 and Exp2, left and right panels, respectively) for the enrichment of SINV-derived small RNA in Piwi4-IP fraction compared to input sample. (B) Lack of enrichment of SINV-derived small RNA in control eGFP immunoprecipitation (IP) fraction compared to input sample. (C) Network of Piwi4 protein interactions identified by affinity purification followed by mass-spectrometry using a C-terminal FLAG tagged-Piwi4 expressed in Aag2 cells as bait. Proteins known to be involved in piRNA pathways or in antiviral response in insects are shown in green or blue, respectively.
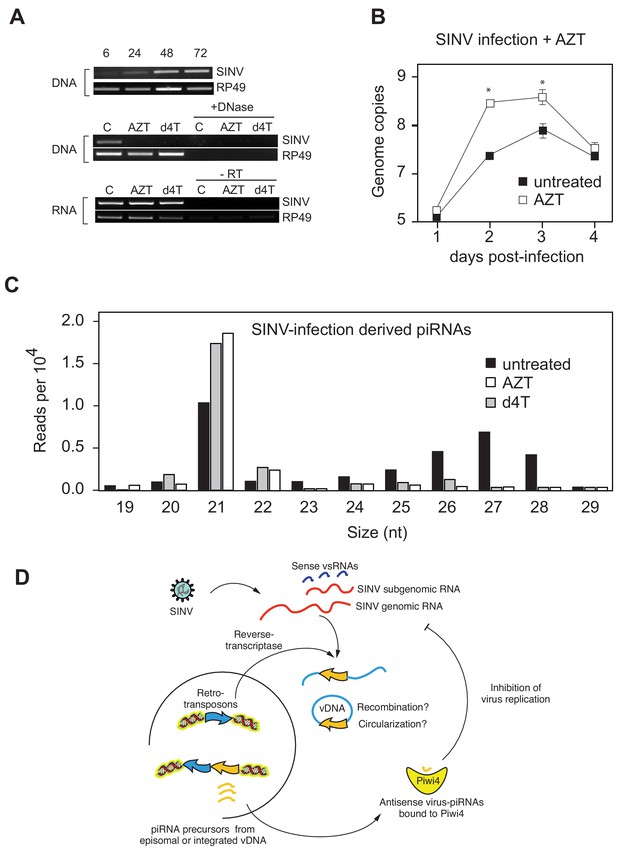
Production of v-piRNAs is blocked by reverse-transcriptase inhibitors.
(A) Multi step growth curve of SINV RNA in Aag2 cells (MOI 0.1) with or without AZT treatment measured by RT-qPCR. RP49 was used as a normalization control. The error bars represent standard deviation of four biological replicates. Significant changes over untreated control are marked with asterisks (p<0.05, Mann-Whitney U test). B Size distribution plot of small RNAs mapping to the SINV genome from untreated infected Aag2 cells or cells treated with the RT-inhibitors AZT or d4T. Read counts per hundred were normalized to SINV genome copy numbers. (C) Schematic representation of the v-piRNA production. vDNA localized in the nucleus where it is transcribed to produce primary piRNA transcripts that are processed by and bound to Piwi4, and transported to the cytoplasm where they participate in the targeting of viral RNA.
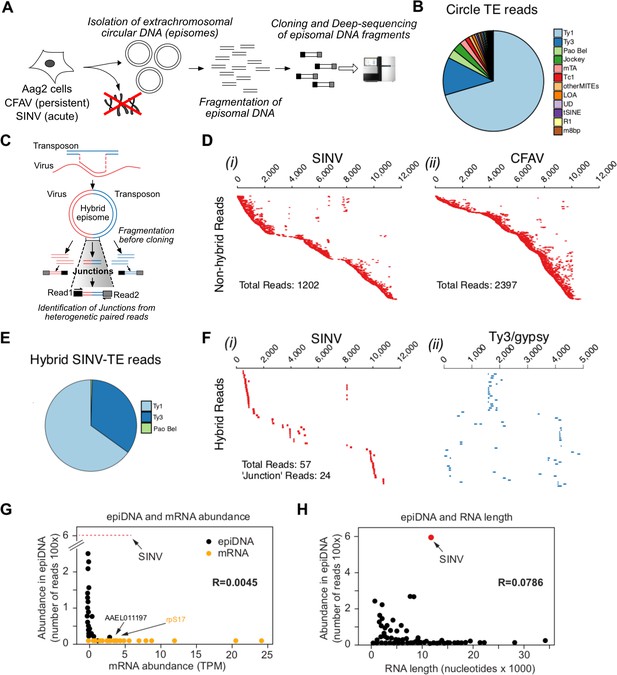
Conversion of viral RNA genome into viral DNA by recombination with retrotransposons is a discriminatory process.
(A Protocol for circular DNA sequencing extracted from infected cells. DNA was extracted from virus-infected Aag2 cells, circular DNA was purified by standard plasmid isolation protocol, treated with exonucleases and cloned for deep-sequencing using Nextera Illumina protocol. (B) Distribution of transposon (TE)-mapping reads from circular DNA sequencing. Of the 12 most abundant transposon families, all were retrotransposons except Tc1 and MITES/m8bp. (C Schematic representation of potential LTR-transposon/virus recombinants and the expected result in the circular DNA population. If virus/transposon recombinants accumulate during infection, hybrid reads should be present. In this example, paired-end read sequences should reveal viral sequences from read one and transposon sequence from read 2. D) All non-hybrid paired-end reads corresponding to SINV (i) or CFAV (ii) are represented by dashes aligned with respect to the corresponding genome (ordered vertically by most 5’-end sequence). Total number of reads observed varied from 1202 to 2397 depending on the virus. Length of dashes corresponds to length of read. Paired reads are shown on the same line. The respective genome lengths of SINV and CFAV are 11,703 and 10,695 nt. E Distribution of SINV-transposon hybrids reads from circular DNA sequencing. All transposon elements recombined with SINV sequences belong to the 3 families of LTR transposons. F Hybrid paired-end reads between SINV and Ty3/gypsy element 73 in SINV infected Aag2 cells. Viral-mapping (blue dashes i), and transposon-mapping (red dashes, ii) from paired read sequences are shown on the same line (across both virus and transposon). Reads are ordered based on the alignment to the CFAV viral genome starting with the most 5’-end sequence. ‘Junction’ reads refer to any paired-end reads where at least one read contained both virus and transposon sequence. G) Distribution of mRNA expressed in Aag2 cells based on their relative expression levels (in Transcripts Per Million, TPM) and their abundance in episomal DNA (in Nextera read counts). Aag2 mRNA with or without sequences identified in episomal DNA are shown in black and yellow, respectively. SINV abundance in episomal DNA is shown as a red line. The correlation coefficient R is shown. (H Distribution of mRNA expressed in Aag2 cells based on their length and abundance in episomal DNA (in Nextera read counts). The correlation coefficient R is shown.
-
Figure 5—source data 1
Identification of virus-derived episomal DNAs in Sindbis or Dengue virus infected Aag2 cells.
Aag2 cells were infected with Sindbis or Dengue virus and collected 3 days after infection and processed DNA extraction (Nucleospin tissue kit, Macherey-Nigel). DNA samples were treated with Plasmid-Safe exonuclease to remove non-circular DNA.
- https://doi.org/10.7554/eLife.41244.014
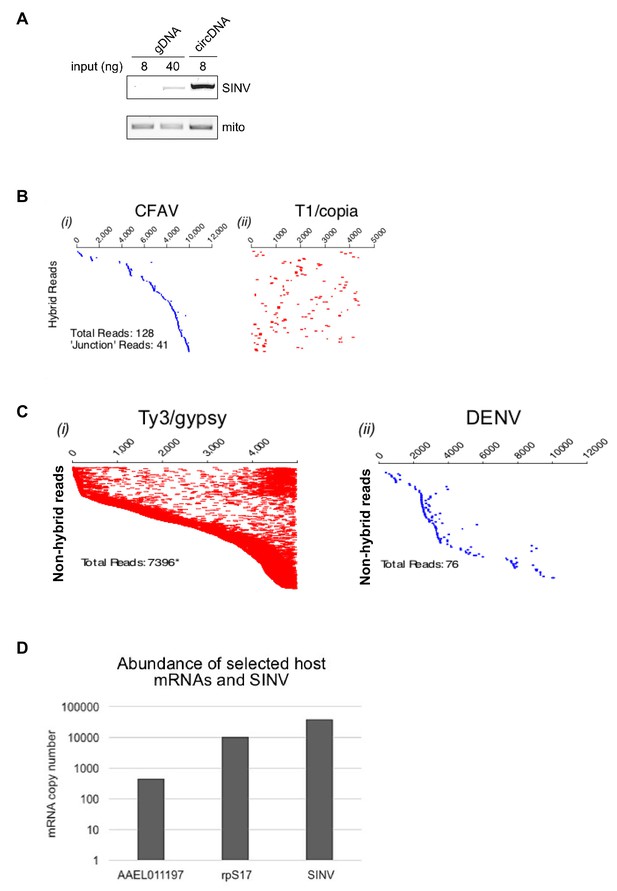
Analysis of episomal viral and transposon-derived DNA in arbovirus-infected Aag2 cells.
(A) SINV infected-Aag2 cells were collected at 4 d.p.i and processed for genomic DNA (gDNA) or episomal circular DNA (circDNA) extraction. 8 or 40 ng gDNA, and 8 ng circDNA were used to detect SINV derived vDNA by end point PCR. Mitochondrial DNA was detected in all sample, with circDNA only showing mild enrichment of it, likely due to the limited efficiency of the plasmid prep kit (used for circDNA isolation) in extracting genetic material from mitochondria. (B) Hybrid paired-end reads between CFAV and Ty1/copia element 56. Viral- mapping (blue dashes, (i) and transposon-mapping (red dashes, ii) paired read sequences are shown on the same line (across both virus and transposon). Reads are ordered based on the alignment to the CFAV viral genome starting with the most 5’-end sequence. ‘Junction’ reads refer to any paired-end reads where at least one read contained both virus and transposon sequence. (C) Coverage of non-hybrid paired-end reads corresponding to (i) Ty3/gypsy or (ii) DENV (ordered vertically by most 5’-end sequence). The respective genome lengths of Ty3/gypsy Ele73 and DENV are 5131 and 10,700 nt.( D) Absolute quantification of SINV RNA levels and of two selected host mRNAs found (AAEL011197) or not (rpS17) in episomal DNA in SINV infected Aag2 cells.
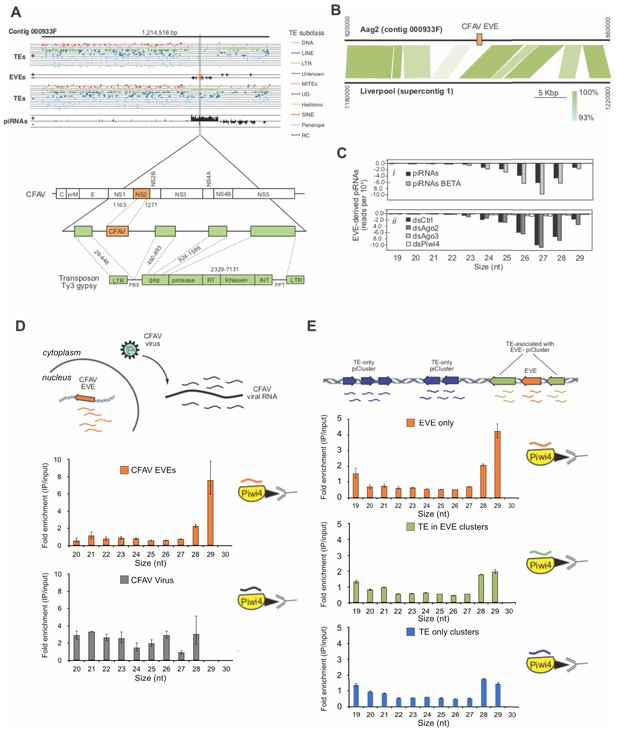
Acquisition of a CFAV EVE in the Aag2 genome and specific association of CFAV EVE-piRNAs with Piwi4.
A (Upper panel) Distribution of transposons (TEs) and CFAV-derived EVE sequences and their respective mapping piRNAs and orientation within the genome (sense +, antisense -) in contig 933F from Aag2 genome. (Lower panel) Organization of the piRNA cluster containing the endogenized CFAV genome region (NS2) within Ty3/gypsy sequences. B Comparison of contig 933F in Aag2 genome and supercontig one in Liverpool mosquito genome. CFAV-derived EVE (orange) is only present in the interspersed transposon (green) sequences in the Aag2 genome. Sequence identity between the two contigs is expressed by color intensity according to the green scale. (C) Characterization of EVE-derived piRNAs (i) Size distribution plot of small RNAs mapping to the EVEs from Aag2 cells with or without β-elimination treatment. (ii) Size distribution plot of small RNAs mapping to the EVEs from Aag2 cells treated with control, Ago2, Piwi4, or Ago3 dsRNA. (D) Enrichment of antisense CFAV EVE (upper panel) or virus (lower panel)-derived small RNA in Piwi4-immunoprecipitation (IP) fraction compared to input sample. (E) Enrichment of genome-wide antisense EVEs (upper panel), TEs associated with EVE-piRNA clusters (middle panel) or TE-only piRNA clusters (lower panel) -derived small RNA in Piwi4-immunoprecipitation (IP) fraction compared to input sample.
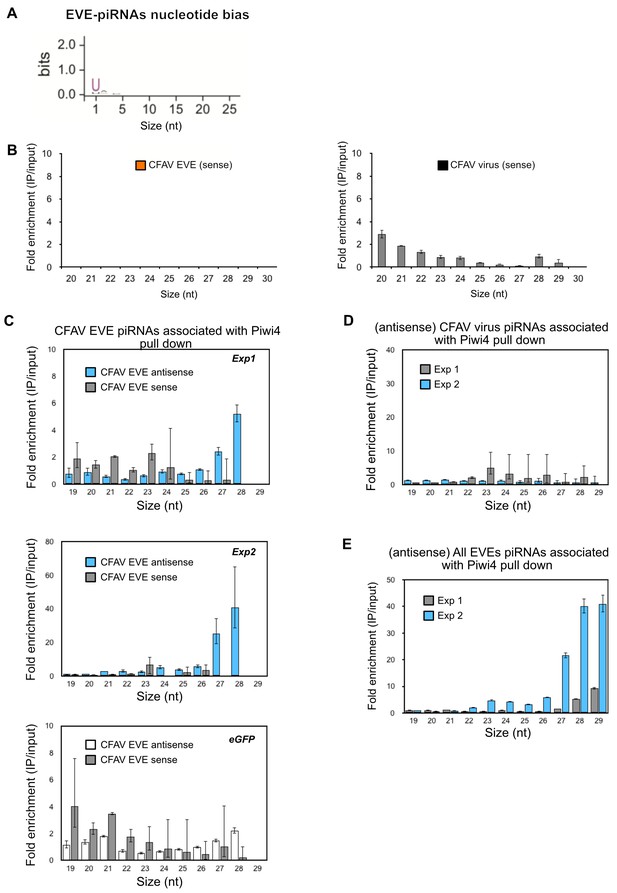
Piwi4 binds to bona-fide antisense v-piRNAs produced by genomic CFAV-EVEs.
(A) The base bias for each position of EVE-derived piRNAs co- immunoprecipitated with Piwi4-FLAG shows a Uracil bias at position 1, characteristic of antisense Piwi-associated piRNAs (shown in bits). (B) Lack of enrichment of sense CFAV EVE (left panel) or virus (right panel)- derived small RNA in Piwi4-IP fraction compared to input sample. (C) Confirmation of specific enrichment for CFAV EVE-derived antisense piRNAs Piwi4-IP fraction compared to input sample in two additional independent experiments (Exp1 and Exp2, two upper panels). The mock pull down experiment (eGFP) did not show enrichment for antisense piRNAs derived from CFAV EVE. (D) Lack of enrichment of antisense CFAV virus derived small RNAs in Piwi4-IP fraction compared to input sample in the tow additional Piwi4 pull-down experiments. (E) Confirmation of specific enrichment for EVE-derived antisense piRNAs Piwi4-IP fraction at the genome-wide level compared to input sample in two additional independent experiments (Exp1 and Exp2).
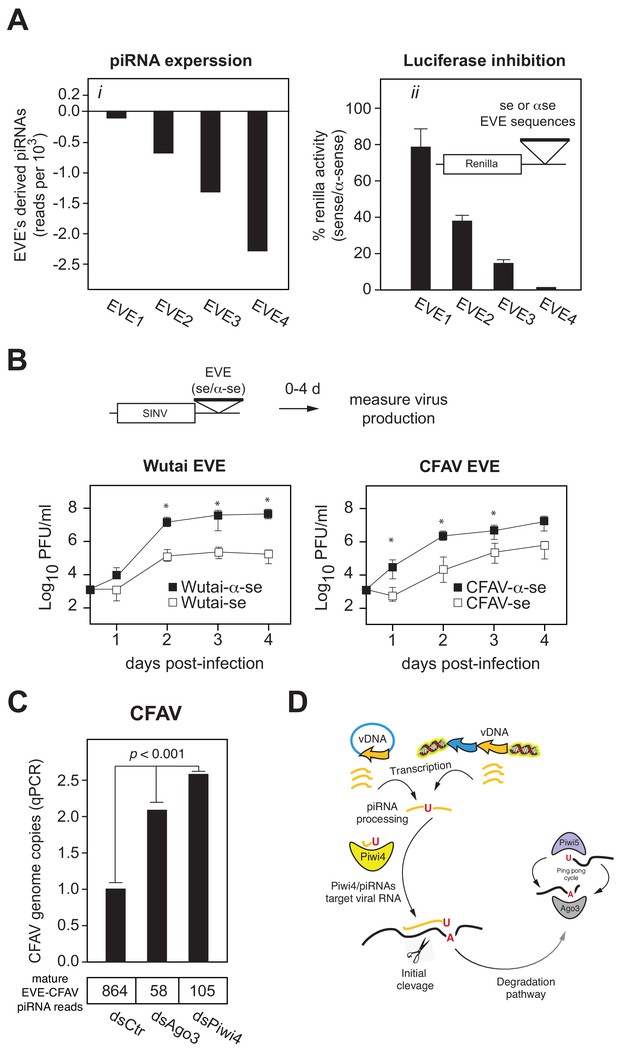
EVEs produce functional antiviral piRNAs that regulate virus attenuation.
(A) (i) Abundance of piRNA reads (24-30nt long) derived from four distinct EVEs compared to the total number of small RNAs isolated from Aag2 cells (in reads per thousand). (ii) EVE-piRNA silencing reporters were designed by cloning a ~ 500 bp sense or antisense sequence from the identified piRNA producing EVEs in (i) into the 3’ UTR of a Renilla luciferase expression vector (see schematic in inset). Silencing activity was measured as the ratio of Renilla luciferase activities from sense versus antisense EVE targets (normalized to firefly luciferase activity). (B) EVE-piRNA antiviral activity was assessed by infecting Aag2 cells with four modified SINV strains containing a ~ 350 nt sense or antisense sequence from identified piRNA-producing EVEs, targeting either Wutai Mosquito virus (left panel) or CFAV (right panel). Replication of SINV strains with a sense target to the endogenously expressed cognate piRNAs was significantly reduced compared to their antisense counterparts (asterisks correspond to p<0.05, one tailed Mann-Whitney-Wilcoxon test, n = 3 independent experiments). (C) Aag2 were transfected with dsRNA control (dsCtr), or directed against Ago3 (dsAgo3) or Piwi4 (dsPiwi4) and copies of CFAV genome per cell were determined by qPCR. Bottom table shows the number of mature piRNAs (i.e.: beta-elimination resistant) per condition in reads per millions.
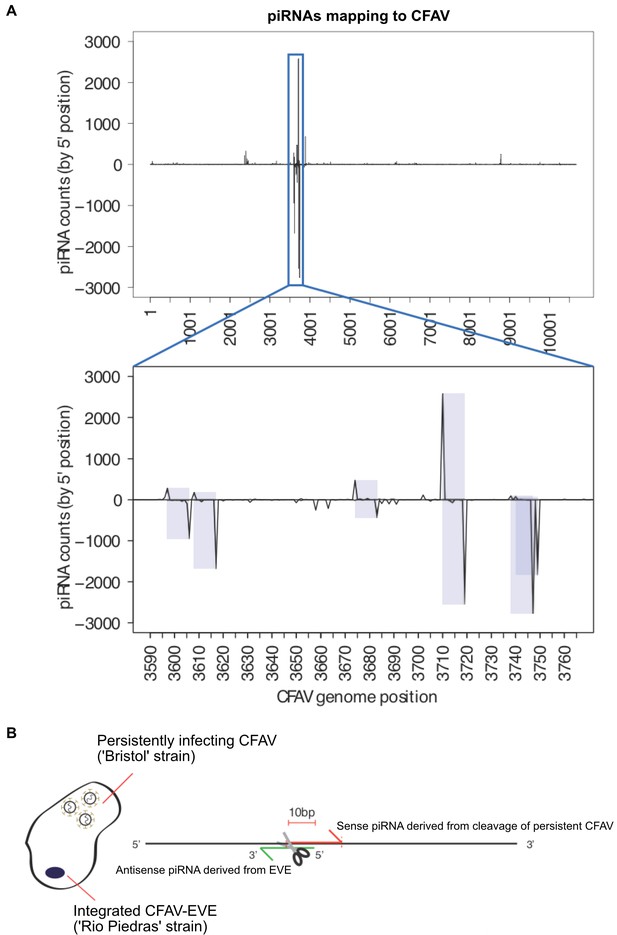
piRNA mapping to CFAV viral genome and EVEs reveals multiple signatures of CFAV virus targeting by EVE-derived piRNAs.
(A) Identification of multiple sense/antisense CFAV piRNAs pairs showing typical ping-pong, 10 base offset signatures.(B) Schematic of the relationship between an antisense CFAV EVE derived piRNA and its sense CFAV virus piRNA counterpart.
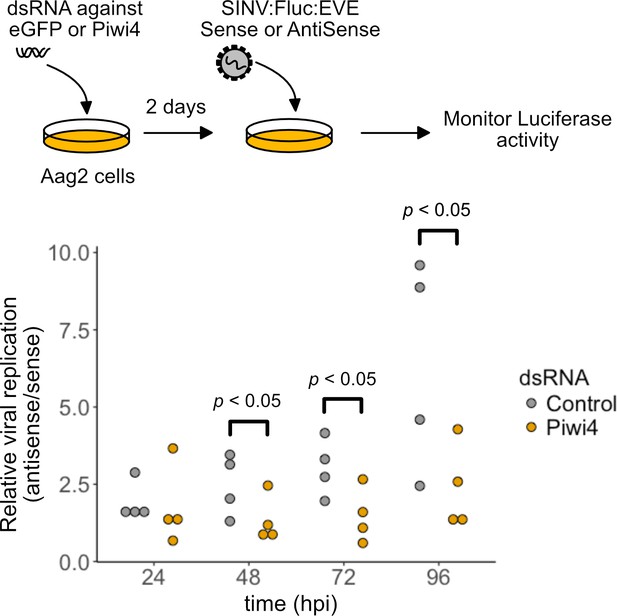
EVE-piRNA antiviral activity is mediated by Piwi4.
Aag2 cells, transfected with dsRNA directed against Piwi4 or eGFP, were infected with a modified SINV strains (SINV:Fluc:EVE) expressing the Firefly luciferase reporter and carrying a 120 nt sense (S) or antisense (AS) sequence from an identified piRNA-producing EVE inserted within its 3’ UTR. Specific inhibition of the sense EVE target containing SINV:Fluc:EVE was assessed as the luciferase activity ratio of the SINV with the AS EVE over S EVE, at 24, 48, 72 and 96 hr post infection (hpi). The relative viral replication advantage of the AS vs S EVE containing virus was reduced in Piwi4 knock-down in Aag2 (Wilcoxon Signed-Rank test, n = 4 independent experiments).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Aedes aegypti) | Ago3 | https://www.vectorbase.org | Previously known as LOC5569680 | |
| Gene (Aedes aegypti) | Piwi4 | https://www.vectorbase.org | Previously known as AAEL007698 | |
| Gene (Aedes aegypti) | Ago2 | https://www.vectorbase.org | Previously known as AAEL017251 | |
| Strain, strain background (Sindbis virus) | Sindbis virus | Strauss et al., 1984 | Sindbis virus was produced and titered in BHK-21 cells. | |
| Strain, strain background (Dengue virus) | Dengue Virus type 2 Thailand 16681 | Kinney et al., 1997 | ||
| Strain, strain background (Dengue virus) | Dengue Virus type 2 Jamaica 1409 | Pierro et al., 2006 | ||
| Strain, strain background (Aedes aegypti) | Chetumal strain | NA | Ae. aegypti female mosquitoes were reared, infected and injected in the Olson lab | |
| Transfected construct (Ae. Aegypt) | pAcFLuc | This paper | Ae. Aegypti poly-ubiquitin promoter driving expression of Firefly Luciferase | |
| Transfected construct (D. melanogaster) | pUbRLuc | This paper | D. melanogaster Actin promoter driving expression of Renilla Luciferase | |
| Transfected construct (Ae. Aegypi) | pUb:FLAG-Piwi4 | This paper | Ae. Aegypti poly-ubiquitin promoter driving expression of a FLAGx3-tagged Piwi4 coding sequence | |
| Antibody | anti-FLAG M2 Magnetic beads (mouse monoclonal) | Sigma-Aldrich | M8823 | |
| Recombinant DNA reagent | SINV:Fluc-EVE-Sense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter to express Firefly luciferase with a 185nt sense Xincheng mosquito virus glycoprotein EVE fragment in its 3'UTR. | |
| Recombinant DNA reagent | SINV:Fluc-EVE-AntiSense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter to express Firefly luciferase with the 185nt sense Xincheng mosquito virus glycoprotein EVE fragment in the antisense orientation in its 3'UTR. | |
| Recombinant DNA reagent | SINV:Wutai-Sense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter followed by a 334nt sense Wutai nucleocapsid EVE fragment in its 3'UTR. | |
| Recombinant DNA reagent | SINV:Wutai-AntiSense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter followed by the 334nt Wutai nucleocapsid EVE fragment in antisense orientation in its 3'UTR. | |
| Recombinant DNA reagent | SINV:CFAV-Sense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter followed by a 326 nt sense CFAV NS2a EVE fragment in its 3'UTR. | |
| Recombinant DNA reagent | SINV:CFAV-AntiSense | This paper | Sindbis virus strain modified to include an additional subgenomic promoter followed by the 326nt CFAV NS2a EVE fragment in antisense orientation in its 3'UTR. | |
| Commercial assay or kit | Luciferase Assay System | Promega | E1500 | |
| Commercial assay or kit | Dual-Glo Luciferase Assay System | Promega | E1910 | |
| Commercial assay or kit | NEBNext Small RNA Library Prep Set for Illumina | NEB | E7330S | |
| Chemical compound, drug | 3'-azido-3'-deoxythymidine (AZT) | Synthonix | A2698 | |
| Software, algorithm | Bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml | |
| Software, algorithm | Python (version 2.7.6) | Python Software Foundation,; 2017 | https://www.python.org | |
| Software, algorithm | R (version 3.30) | R Core TeamR: a language and environment for statistical computing. R Foundation for Statitstical Computing; 2014 | https://r-project.org/ | |
| Software, algorithm | FASTX Toolkit | Hannon lab | http://hannonlab.cshl.edu/fastx_toolkit/ | |
| Software, algorithm | WebLogo 3 | Crooks et al., 2004 | weblogo.threeplusone.com |
Number of total reads for virus and TE derrived piRNAs.
https://doi.org/10.7554/eLife.41244.020| Normal | dsCtr | dsAgo2 | dsAgo3 | dsPiwi4 |
|---|---|---|---|---|
| Total reads | 14779383 | 13697808 | 10207667 | 12139804 |
| virus | 194682 | 659432 | 125859 | 301905 |
| TE | 1617084 | 1220933 | 782789 | 1484997 |
Number of reads for virus and TE derrived piRNAs following beta elimindation.
https://doi.org/10.7554/eLife.41244.021| Beta | dsCtr-B | dsAgo2-B | dsAgo3-B | dsPiwi4-B |
|---|---|---|---|---|
| Total reads | 14008617 | 16135731 | 6153485 | 10108849 |
| virus | 242815 | 562820 | 38657 | 23246 |
| TE | 2109794 | 2163585 | 250759 | 331526 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41244.022





