Intramolecular domain dynamics regulate synaptic MAGUK protein interactions
Figures
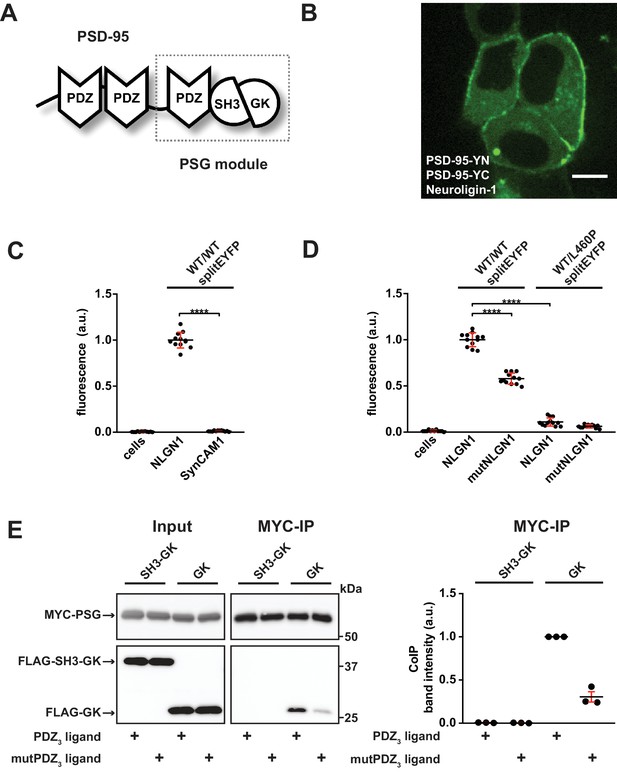
PDZ3 ligand-induced dynamics in the PDZ3-SH3-GK module facilitate oligomerisation.
(A) Schematic representation of the PSD-95 domain organisation. PSD-95 contains three PDZ domains followed by a SH3-GK domain tandem. The PSG module (PDZ3-SH3-GK) is common to the MAGUK protein family. (B) Live-cell microscopy of HEK-293T cells transfected with PSD-95-YN, PSD-95-YC and full-length Neuroligin-1 reveals a membrane associated localisation of the refolded complex (transfection corresponding to WT/WTsplitEYFP plus NLGN1 in Figure 1C,D). Scale bar: 10 µm. (C, D) PSD-95 oligomerisation assay based on BiFC. HEK-293T cells were triple-transfected with the displayed DNA constructs and EYFP refolding was assessed by flow cytometry. Formation of oligomeric fluorescent complexes is effective in the presence of wild-type Neuroligin-1 (NLGN1). (C) Fluorescence is almost not detectable by coexpression of SynCAM1 (SynCAM1 is not binding to PSD-95 PDZ domains) (D) Fluorescence is reduced by either site-directed mutagenesis of the NLGN1 PDZ3 ligand C- terminus (mutNLGN1: TTRV ► TARA), or a targeted amino acid exchange in the PSD-95 SH3 domain (L460P). (C, D) The dot plots indicate mean values (black horizontal bar) with SD (red vertical bar), based on twelve individual measurements (dots) that originate from four independent experiments (results from each experiment are triplicates for each DNA construct combination). Data were analysed by one-way ANOVA/Sidak's multiple comparisons test. ****p<0.0001. (E) MYC-PSG and FLAG-SH3-GK or FLAG-GK were coexpressed together with either CRIPT-derived PDZ3 or mutPDZ3 ligand constructs. Upon MYC-PSG IP, proteins were analysed by western blot with αFLAG antibodies. Coexpression of the CRIPT-derived PDZ3 ligand enhanced the coIP of PSG and GK, whereas coIP of PSG and SH3-GK was negligible regardless of whether or not the CRIPT-derived PDZ3 ligand construct was coexpressed. The western blot shown (left side) is a representative example of three independent experiments; the corresponding quantification of coIP band intensities from these three experiments is shown in the dot plot on the right side indicating mean values ± SEM.
-
Figure 1—source data 1
Source data for Figure 1C,D.
- https://doi.org/10.7554/eLife.41299.003
-
Figure 1—source data 2
Source data for Figure 1E.
- https://doi.org/10.7554/eLife.41299.004
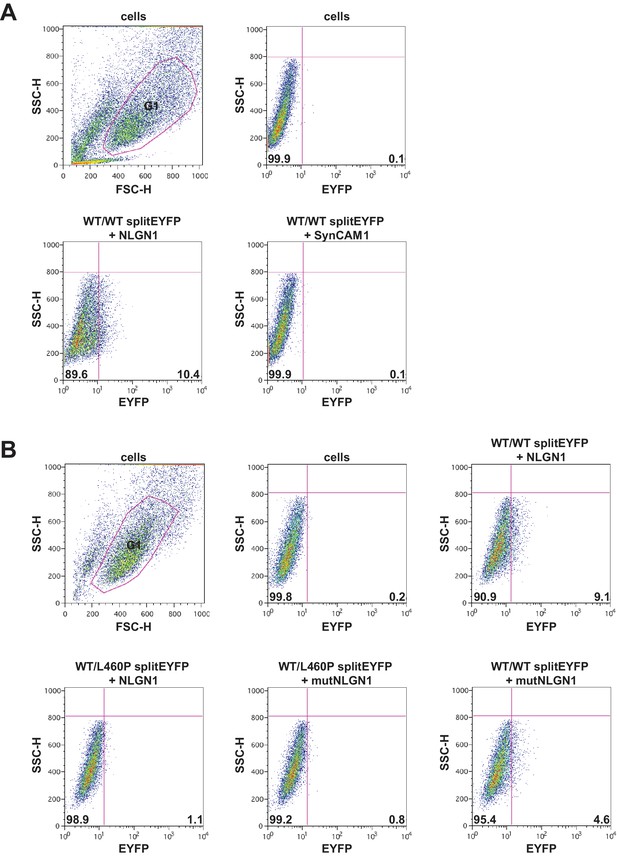
FACS plots for Figure 1C,D.
(A, B) Gating strategy and representative dot plots of the PSD-95 oligomerisation assay as shown in Figure 1C,D. Untransfected cells or cells transfected with the indicated constructs were harvested and analysed by flow cytometry. The HEK-293T cell population was defined by the gate G1 in the forward scatter height (FSC-H) versus side scatter height (SSC-H) plot. (A and B upper left panel). 10,000 cells from the gate G1 were then subsequently analysed by plotting side scatter height (SSC-H) versus yellow fluorescence (EYFP: enhanced yellow fluorescent protein) emitted by the refolded splitEYFP halves. Fluorescent cells appear as dots in the lower right quadrants.
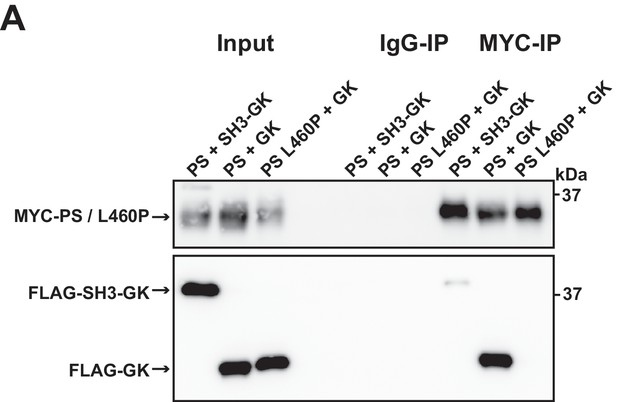
Supplement for Figure 1D.
(A) PSD-95 constructs consisting of the PDZ3-SH3 domains (PS) were coexpressed together with either an SH3-GK domain construct, or a GK domain construct. As a comparison PDZ3-SH3 L460P was coexpressed with a GK domain construct and PDZ3-SH3/PDZ3-SH3 L460P constructs were precipitated and copurified proteins were identified by western blot. By mutating the leucine 460 to proline this efficient protein complex formation is disrupted. By exchanging the internal L460 residue the SH3 domain loses its ability to bind to the GK domain construct in trans.
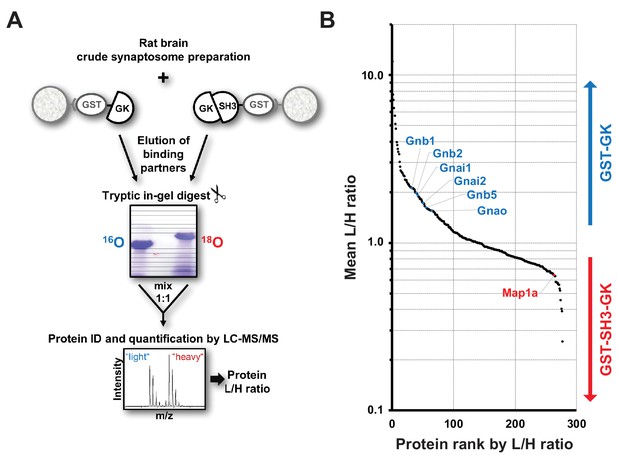
Identification of interactors that differentially bind to PSD-95 C-terminal domains.
(A) Schematic representation of the quantitative mass spectrometry experiment to identify PSD-95 GK domain interactors from crude rat synaptosomes by GST pull-down of bacterially expressed GST-GK or GST-SH3-GK constructs and 18O-labeling. (B) GST pull-downs were performed in triplicates and 278 interacting proteins that passed our threshold settings were identified and quantified by mass spectrometry. Proteins are ranked by their mean L/H ratio indicating preferential enrichment with either GST-GK or GST-SH3-GK constructs. The heterotrimeric G protein subunit Gnb5 was found to be enriched in the GST-GK fraction relative to the GST-SH3-GK fraction and selected for further studies.
-
Figure 2—source data 1
Source data for Figure 2B.
- https://doi.org/10.7554/eLife.41299.008
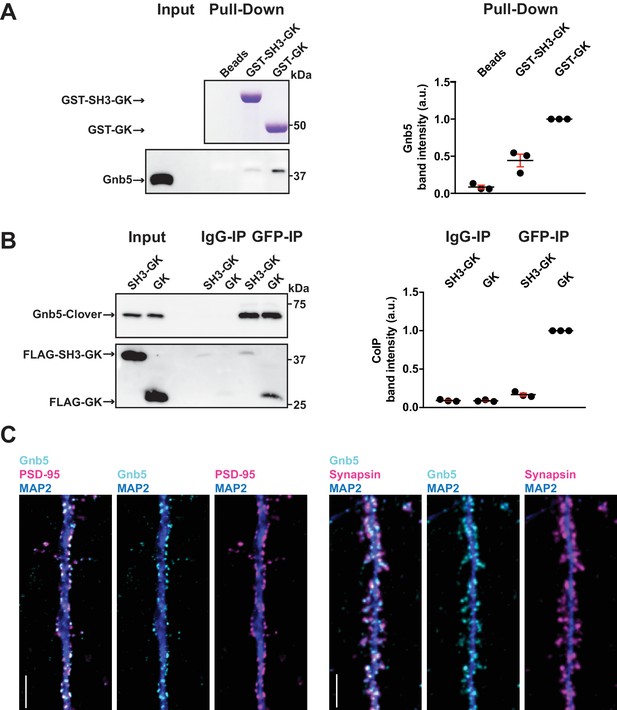
The heterotrimeric G protein subunit Gnb5 is a novel PSD-95 interactor.
(A) GST pull-down from crude synaptosomal proteins (comparable amounts of GST tagged proteins observable by Coomassie, upper panel) enabled comparison of Gnb5 binding to the GK domain alone versus the SH3-GK domain. Gnb5 is effectively enriched in the GST-GK pull-down compared to bead controls or GST-SH3-GK pull-downs, as observed by western blot with a commercially available αGnb5 antibody (lower panel). The GST pull-down shown on the left side is a representative example of three independent experiments; the corresponding quantification of copurified Gnb5 band intensities from these three experiments is shown in the dot plot on the right side indicating mean values ± SEM. (B) CoIP experiment of tagged Gnb5 (Gnb5-Clover) with tagged SH3-GK or GK (FLAG-SH3-GK or FLAG-GK). Immunoprecipitation of Gnb5-Clover with αGFP antibody efficiently copurified the GK-domain construct (observed via western blot with αFLAG antibodies, lower panel). The western blot shown (left side) is a representative example of three independent experiments; the corresponding quantification of coIP band intensities from these three experiments is shown in the dot plot on the right side indicating mean values ± SEM. (C) Cultures of rat hippocampal neurons (E18) were fixed at DIV21 and stained for Gnb5 together with the dendritic marker MAP2 (microtubule-associated protein 2) and either the postsynaptic protein PSD-95 (left panel) or the presynaptic marker Synapsin (right panel) and respective fluorescent secondary antibodies, and visualised by confocal microscopy. Scale bars: 5 μm.
-
Figure 3—source data 1
Source data for Figure 3A,B.
- https://doi.org/10.7554/eLife.41299.010
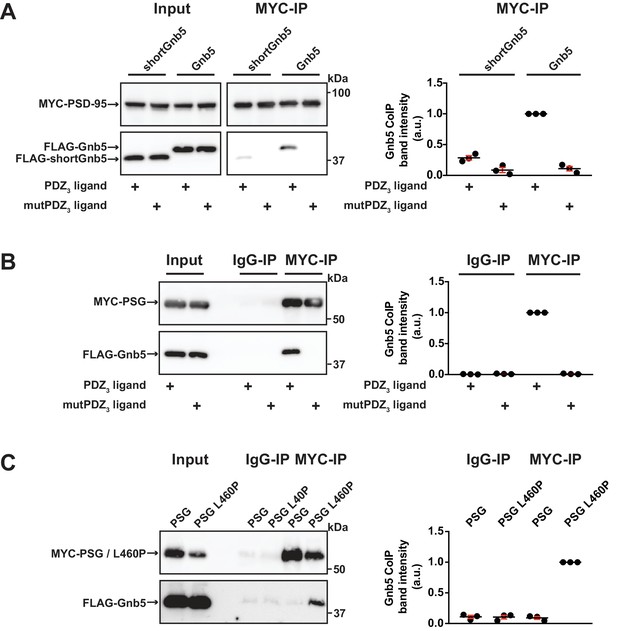
Gnb5 - PSD-95 complex formation is regulated by CRIPT-derived PDZ3 ligand binding.
For this figure, western blots shown on the left side are representative examples of three independent experiments; the corresponding quantification of coIP band intensities from these three experiments are shown in the dot plots on the right side indicating mean values ± SEM. (A) MYC-PSD-95 and FLAG-Gnb5 or FLAG-shortGnb5 were coexpressed with either CRIPT-derived PDZ3 or mutPDZ3 ligand constructs. MYC-PSD-95 was precipitated and proteins were analysed by western blot with αFLAG antibodies. Coexpression of the CRIPT-derived PDZ3 ligand facilitated the coIP of PSD-95 and Gnb5, coIP with the shortGnb5 construct (N-terminal truncation) was much less efficient. In the presence of the CRIPT-derived mutPDZ3 ligand, coprecipitated proteins were not detectable. (B) CoIP of MYC-PSG and FLAG-Gnb5 together with either CRIPT-derived PDZ3 or mutPDZ3 ligand constructs. The presence of CRIPT-derived PDZ3 ligand constructs facilitated coprecipitation of PSG and Gnb5 (see comparative western blot with αFLAG antibodies, lower panel). (C) Coexpression of MYC-PSG or MYC-PSG L460P with FLAG-Gnb5 and subsequent MYC IP. PSG L460P IP efficiently copurifies Gnb5 (observed by western blot with αFLAG antibodies).
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.41299.012
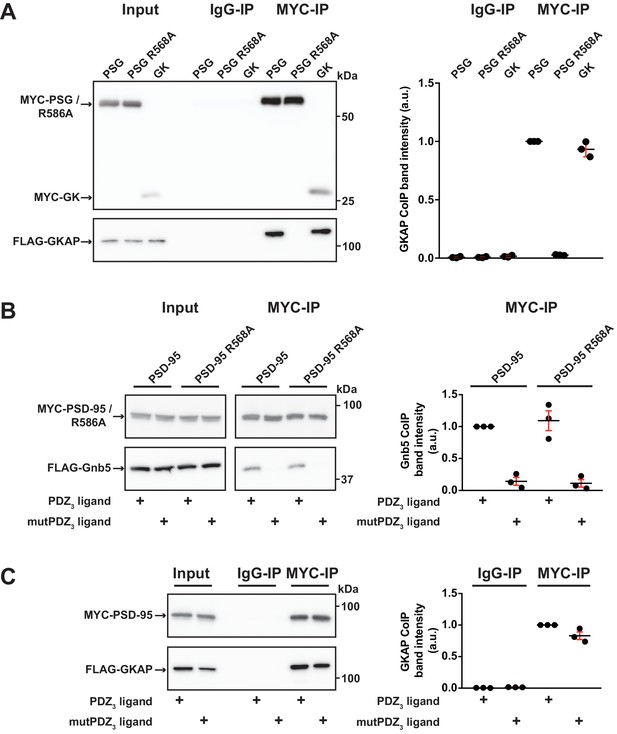
GK domain interactions are differentially regulated.
For this figure, western blots shown on the left side are representative examples of three independent experiments; the corresponding quantification of coIP band intensities from these three experiments are shown in the dot plots on the right side indicating mean values ± SEM. (A) MYC-PSG, MYC-PSG R568A and MYC-GK were coexpressed with FLAG-GKAP. Following MYC IP, precipitated proteins were analysed by western blot. GKAP coprecipitated with PSG and GK domain constructs. The GK domain mutant PSG R568A was not able to bind GKAP. (B) Following coexpression of MYC-PSD-95 or MYC-PSD-95 R568A with FLAG-Gnb5, together with either CRIPT-derived PDZ3 or mutPDZ3 ligand, proteins were precipitated with αMYC-antibody and analysed by western blot. Gnb5 coIP with either PSD-95 or PSD-95 R568A was efficiently promoted by the presence of PDZ3-binding ligand, irrespective of the GK domain mutation R568A. (C) CoIP of MYC-PSD-95 and FLAG-GKAP together with either CRIPT-derived PDZ3 or mutPDZ3 ligand constructs and analysis of precipitated proteins by western blot with antibodies to the corresponding tags. The presence of CRIPT-derived PDZ3 ligands in the lysate had almost negligible effect on PSD-95 GKAP interaction.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.41299.014
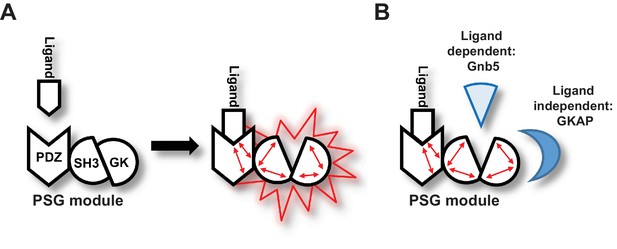
Graphical Summary.
(A) PSD-95 C-terminal domains (PSG module) functionally cooperate and regulate homotypic and heterotypic complex formation. We propose that CRIPT-derived PDZ3 ligand binding to the PDZ3 domain induces a loosening of the intramolecular SH3-GK interaction. This ‘open’ conformation is then able to initiate subsequent oligomerisation and protein binding. (B) Model of CRIPT-derived PDZ3 ligand-dependent and ligand-independent binding to the PSD-95 C-terminal SH3-GK domain tandem. Ligand - PDZ3 domain binding facilitates association with Gnb5.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41299.016





