Pericentrin-mediated SAS-6 recruitment promotes centriole assembly
Figures
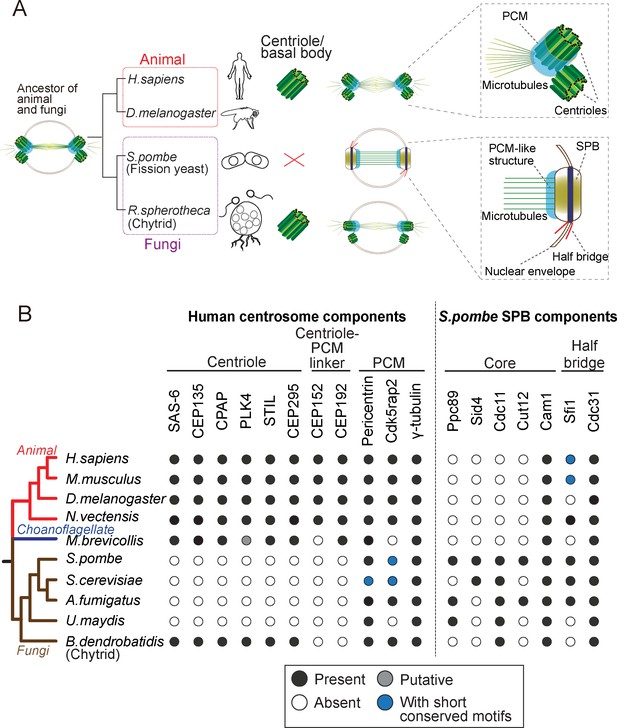
Evolution of the morphology and protein content of animal and fungi centrosomes.
(A) The structure of the centrosome in mitosis in most animals, chytrids (flagellated fungi) and fission yeast. Animals and chytrids have a centriole/basal body and a canonical centrosome composed of a pair of centrioles surrounded by PCM, which anchors and nucleates microtubules. Fission yeast lacks a centriole but has a spindle pole body (SPB) inserted in the nuclear envelope. The SPB nucleates microtubules from the PCM-like structure, inside the nucleus. Parsimoniously, it is likely that the common ancestor of animals and fungi had a centriole-containing centrosome with a PCM structure (model shown). (B) Phylogenetic distribution of centrosome components in opisthokonts (animals, fungi and choanoflagellates). We searched for orthologues of components of the human centrosome localizing to centrioles, centriole-PCM linkers and PCM, and the fission yeast SPB components. Black circles represent the presence of orthologues that were identified by the bidirectional best hit approach to the human or fission yeast proteins, respectively; gray circle represents the presence of a putative orthologue identified by constructing phylogenetic trees; blue circles indicate that previous studies showed the presence of a protein with short conserved motifs (Kilmartin, 2003; Samejima et al., 2010; Lin et al., 2014) although we failed to identify it by the computational methods highlighted above; white circles indicate no detectable orthologue.
-
Figure 1—source data 1
List of the predicted orthologues of the human centrosome and S. pombe SPB components in animal and fungi species.
- https://doi.org/10.7554/eLife.41418.003
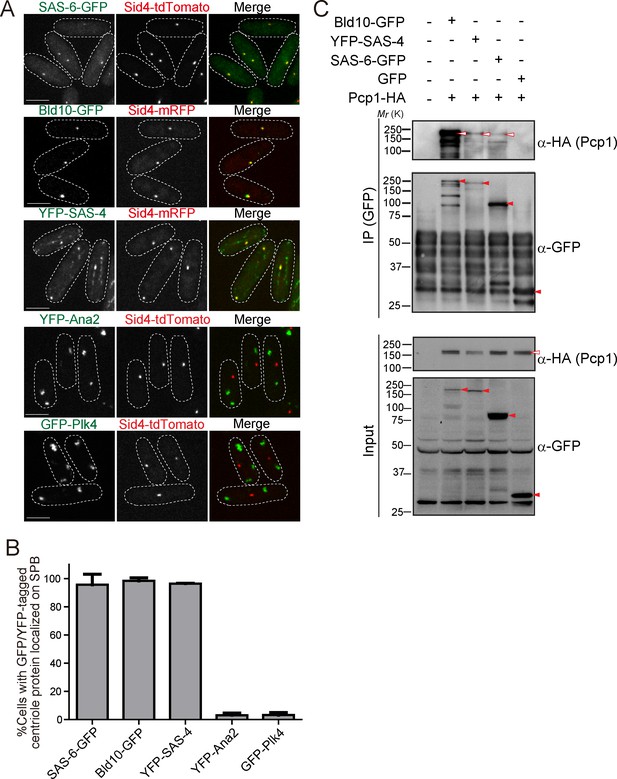
Drosophila centriole proteins localize to the centrosome of fission yeast.
(A) SAS-6, Bld10 and SAS-4 localize to the SPBs, while Ana2 and Plk4 do not (see Materials and methods for details on expression constructs). Scale bar, 5 μm. (B) Quantification of cells with GFP or YFP-tagged centriole proteins localized on the SPB. Data are the average of three experiments ± s.d. (N > 50, GFP/YFP-positive cells). (C) Physical interaction between the centriole proteins and fission yeast Pcp1. Protein extract was prepared from fission yeast cells expressing HA-tagged Pcp1 and either SAS-6-GFP, or Bld10-GFP, or YFP-SAS-4 or GFP. The GFP-tagged proteins were immunoprecipitated with anti-GFP antibody. Immunoprecipitates and inputs (4%) were analyzed by western blotting using the indicated antibodies. Red open and filled arrowheads indicate the bands of Pcp1-HA and GFP/YFP fusion proteins, respectively.
-
Figure 2—source data 1
The source data to plot the graph in Figure 2B.
- https://doi.org/10.7554/eLife.41418.006
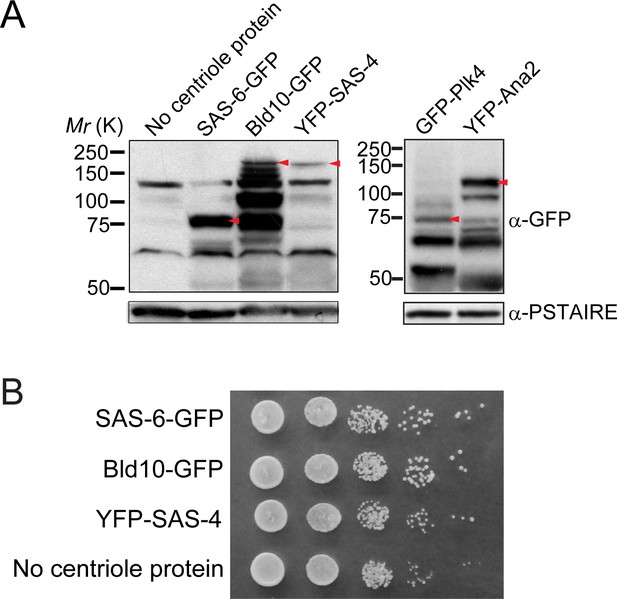
Expression of Drosophila centriole proteins in fission yeast and the growth of fission yeast strains.
(A) Western blotting analysis of the protein extracts prepared from the strains used in Figure 2A. Each red arrowhead indicates the bands with the expected size of the fusion protein. Note that we found that Bld10-GFP was degraded in the cell lysate. It is possible that larger-sized foreign proteins are targeted by fission yeast proteases. (B) Serial dilution assay of the cells expressing indicated Drosophila centriole components under control of the constitutive atb2 promoter, which is shown in Figure 2A. Note that all the strains expressing centriole proteins grew similarly as the control (no centriole protein).
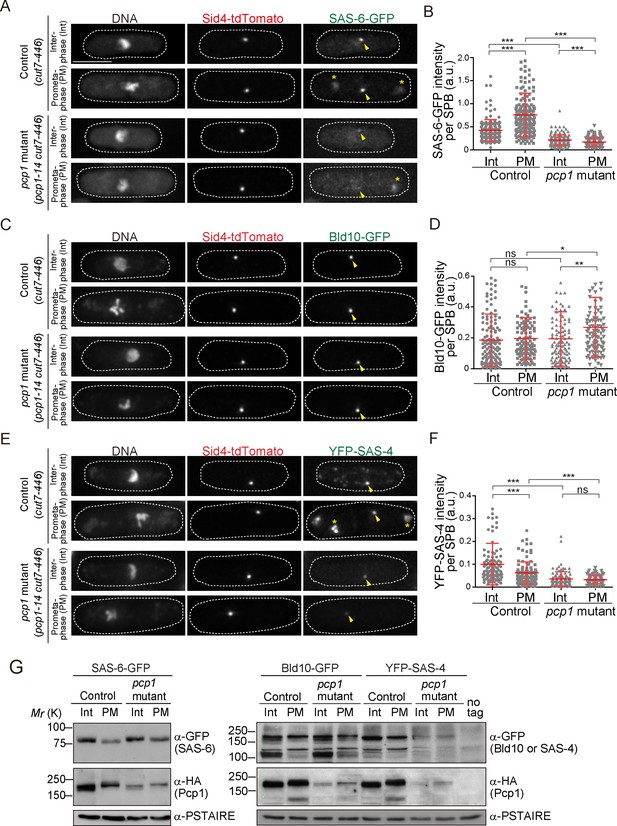
Fission yeast pericentrin-like protein Pcp1 is required to recruit SAS-6 to the SPB.
(A, C, E) SAS-6-GFP, Bld10-GFP and YFP-SAS-4 intensities on the SPB in the pcp1 mutant in asynchronous and prometaphase-arrested cells. The cut7-446 (labeled ‘control’) and cut7-446 pcp1-14 (labeled ‘pcp1 mutant’) strains expressing SAS-6-GFP and Sid4-tdTomato were incubated at the restrictive temperature (36°C) for three hours to block cells in prometaphase due to the cut7-446 mutation (temperature-sensitive allele of a mitotic kinesin, which causes failure in mitotic spindle formation). Representative images of the cells collected before shifting the temperature (interphase, Int) and three hours after the shift to 36°C (prometaphase, PM, restrictive temperature) in control and pcp1 mutant are shown. DNA was stained with DAPI. Arrowheads indicate the signal on the SPB. Note that we also observed aggregation of SAS-6-GFP and YFP-SAS-4 in the cytoplasm both in the control and pcp1 mutant in all the cells exposed to the restrictive temperature (indicated with an asterisk), and never at the normal and permissive culture condition. We think that aggregate formation might stem from the cut7-446 genetic background and/or the heat stress. Scale bar, 5 μm. (B, D, F) Quantification of the intensity of the centriole proteins per SPB in the indicated conditions. Means ± s.d. are shown in red (N > 100 SPBs, ns-not significant, *p<0.05, **p<0.001, ***p<0.0001, Mann-Whitney U test). (G) Western blotting analysis of protein extracts prepared from the indicated conditions.
-
Figure 3—source data 1
The source data to plot the graphs in Figure 3B,D,F.
- https://doi.org/10.7554/eLife.41418.008
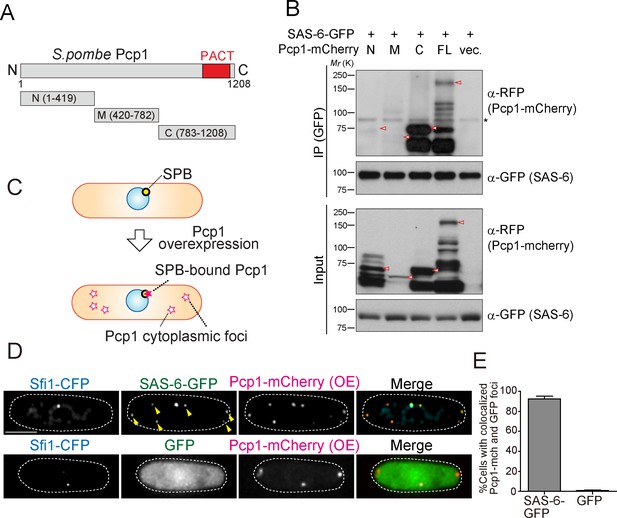
SAS-6 interacts with Pcp1 through the conserved carboxyl-terminal region, and Pcp1 is sufficient for SAS-6 localization.
(A) Schematic illustration of the truncation constructs of Pcp1. (B) Pcp1 interacts with SAS-6 through the conserved carboxyl-terminal region. mCherry-tagged full-length or truncation mutants of Pcp1 were expressed in cells constitutively expressing SAS-6-GFP. Immunoprecipitation was performed and analyzed similarly as in Figure 2B. Red arrowheads indicate bands with the expected size of each fusion protein. The asterisk indicates non-specific bands. (C, D) Pcp1 is sufficient to recruit SAS-6. Overexpression of Pcp1 leads to the formation of Pcp1 containing cytoplasmic foci (schematic illustration, (C). Pcp1-mCherry was overexpressed under control of nmt41 promoter in the strain expressing SAS-6-GFP and GFP alone. Sfi1-CFP is shown (SPB marker). Arrowheads indicate the SAS-6-GFP signal on the Pcp1-cytoplasmic foci. Scale bar, 5 μm. (E) Quantification of the cells with colocalized Pcp1-mCherry and GFP foci. Data are the average of three experiments ± s.d. (N > 50, Pcp1-mCherry positive cells).
-
Figure 4—source data 1
The source data to plot the graph in Figure 4E.
- https://doi.org/10.7554/eLife.41418.016
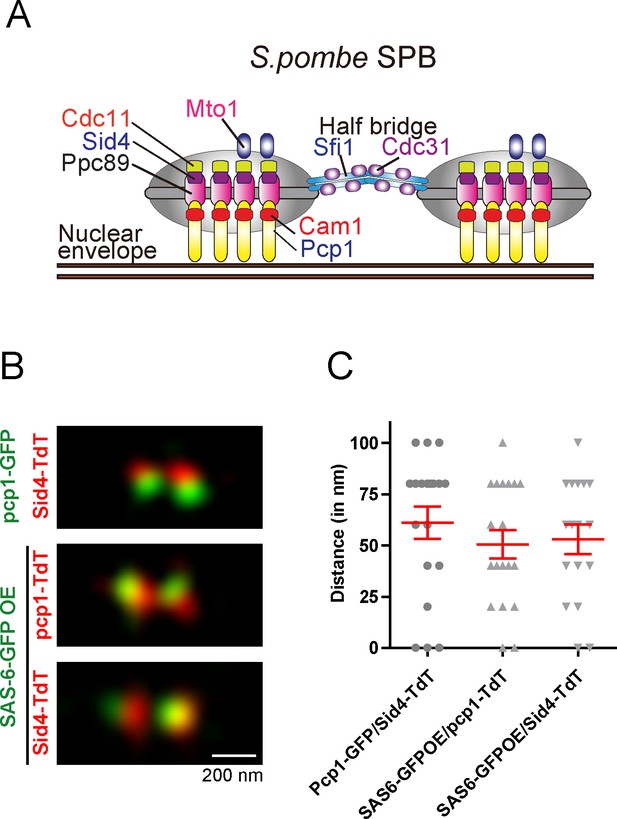
SAS-6-GFP localization within the fission yeast SPB revealed by structured illumination microscopy (SIM).
(A) Schematic representation of localization of components in the duplicated interphase fission yeast SPBs. The illustrated SPB structure was adapted and modified from Ito and Bettencourt-Dias (2018). (B) Representative SIM images of localization of GFP or tdTomato-tagged Pcp1, Sid4 and SAS-6 at the duplicated SPBs. Scale bar, 200 nm. (B) Interfoci distances between the indicated SPB-bound proteins measured by SIM images.
-
Figure 4—figure supplement 1—source data 1
The source data to plot the graph in Figure 4—figure supplement 1C.
- https://doi.org/10.7554/eLife.41418.011
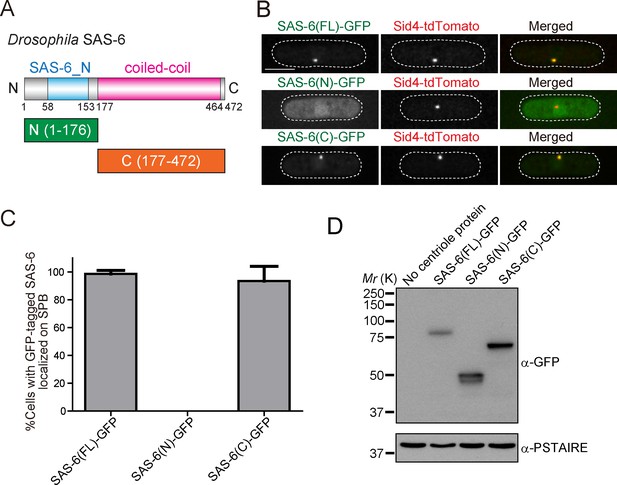
Drosophila SAS-6 localizes to the fission yeast SPB through the C-terminal coiled-coil domain.
(A) Schematic illustration of the truncation constructs of Drosophila SAS-6. (B) Localization of SAS-6 truncation mutants in fission yeast cells. GFP-tagged SAS-6 (full-length (FL), N or C) was expressed under the inducible nmt1 promoter. Expression was induced for 24 hr. Scale bar, 5 μm. (C) Quantification of cells with GFP-tagged full-length or truncated SAS-6 localized at the SPB. Data are the average of three experiments ± s.d. (N > 50, GFP-positive cells). (D) Western blotting analysis of the protein extracts prepared from the strains used in B.
-
Figure 4—figure supplement 2—source data 1
The source data to plot the graph in Figure 4—figure supplement 2C.
- https://doi.org/10.7554/eLife.41418.013
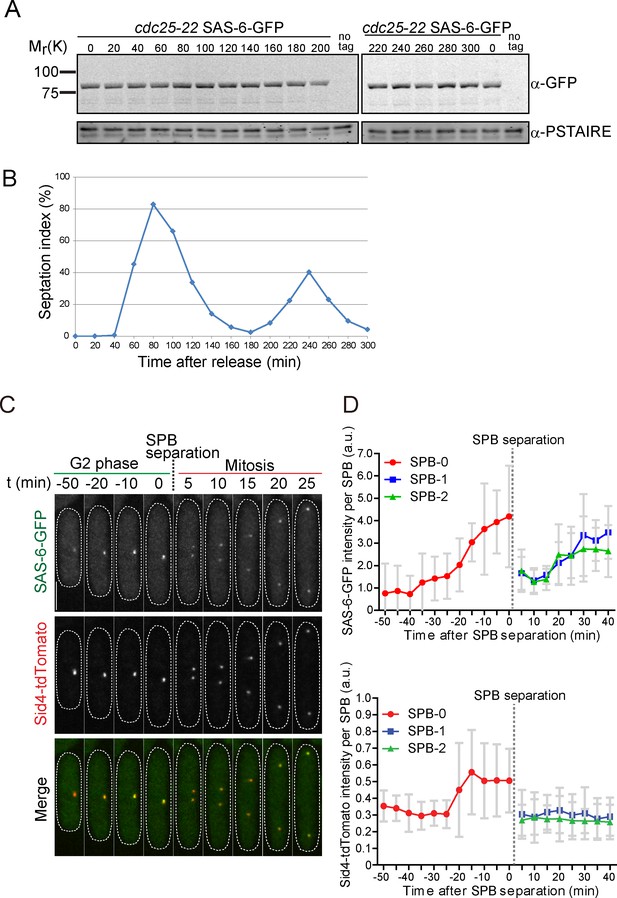
Localization of SAS-6 at the fission yeast SPB is cell-cycle regulated.
(A) The fission yeast strain with cdc25-22 mutation expressing SAS-6-GFP under the atb2 promoter was blocked in the G2/M boundary and synchronously released into mitosis. Cells were collected every 20 min and protein extract was prepared. Protein expression was analyzed by western blotting with anti-GFP and anti-Cdc2 PSTAIRE (loading control). (B) Septation index was determined to check the cell cycle synchronization. Note that cell cycle progression was highly synchronized. (C) SAS-6 localization along the cell cycle. Representative micrographs of time-lapse imaging of cells expressing SAS-6-GFP and Sid4-tdTomato from G2 phase to the end of mitosis. Scale bar, 5 μm. (D) Quantification of the intensity of SAS-6-GFP and Sid4-tdTomato per SPB in the cells shown in C. Mean SAS-6-GFP (top) and Sid4-tdTomato (down) intensity per SPB ±s.d before and after the SPB separation is shown (N = 7). SPB-0, unseparated SPB and SPB-1 and 2, separated SPBs.
-
Figure 4—figure supplement 3—source data 1
The source data to plot the graphs in Figure 4—figure supplement 3B and E.
- https://doi.org/10.7554/eLife.41418.015
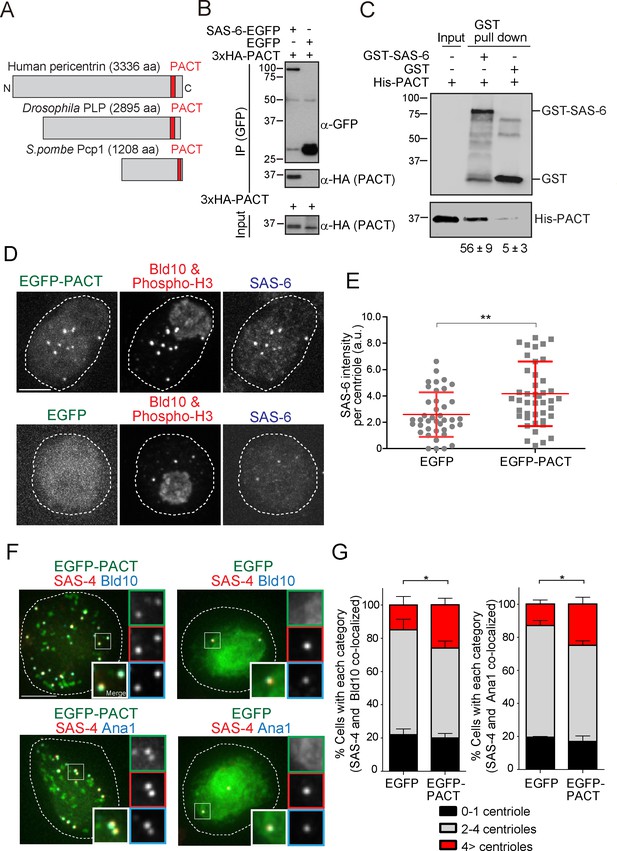
The Drosophila pericentrin (PLP) conserved domain –PACT- interacts with SAS-6, and its overexpression causes centriole amplification.
(A) Schematic illustration of human pericentrin, Drosophila PLP and S. pombe Pcp1. (B) Physical interaction between SAS-6-EGFP and the conserved Drosophila PACT domain. Protein extract was prepared from Drosophila tissue culture cells (D.Mel cells) expressing HA-tagged PACT, and SAS-6-EGFP or EGFP. The GFP-tagged proteins were immunoprecipitated with anti-GFP antibody. Immunoprecipitates and inputs (20%) were analyzed by western blotting using the indicated antibodies. (C) Direct binding between SAS-6 and PACT. The in vitro binding assay was performed using purified GST or GST-fused SAS-6 and His-tagged PLP PACT. Note that we loaded 100% for each sample on each lane after pull-down to compare the efficiency of direct binding (bound vs input). Quantification of His-PACT bound to the GST-fusion protein is shown below the panel (data are the average of three experiments ± s.d) (D) Cells overexpressing EGFP-PACT or EGFP were arrested in mitosis by colchicine treatment for six hours and stained with the antibodies against Bld10 (centriole marker), phospho-H3 (mitotic marker) and SAS-6. Scale bar, 5 μm. (E) Quantification of SAS-6 intensity per centriole in cells overexpressing EGFP-PACT or EGFP arrested in mitosis. Means ± s.d. are shown in red (**p<0.001, Mann-Whitney U test). Results are representative of three independent experiments (N > 40 centrioles for each condition). (F) Cells overexpressing EGFP-PACT or EGFP were stained for two centriole markers (SAS-4 and Bld10, SAS-4 and Ana1) to count centriole number. Scale bar, 5 μm. (G) Quantification of centriole number per cell (N > 50, EGFP-positive cells). Data are the average of three experiments ± s.d. (*p<0.05, Mann-Whitney U test). Note that although control Drosophila tissue culture cells already show cells with underduplicated and over-duplicated centrioles as published before (Bettencourt-Dias et al., 2005), the expression of PACT leads to a significant amplification of centrioles.
-
Figure 5—source data 1
The source data to plot the graphs in Figure 5E and G.
- https://doi.org/10.7554/eLife.41418.018
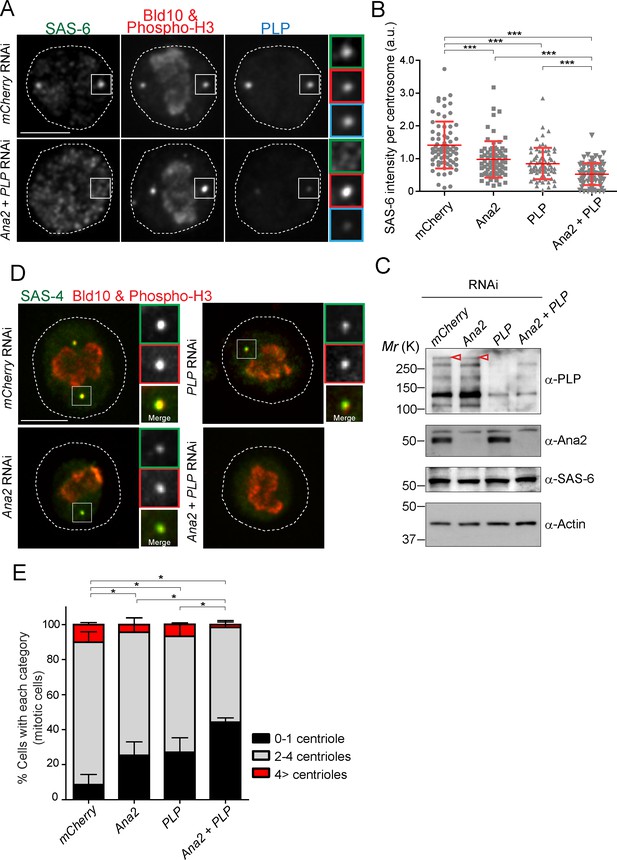
SAS-6-PACT complex formation complements the PLK4-STIL pathway in recruiting SAS-6 to the centrosome, and in promoting centriole biogenesis in tissue cultured cells.
(A) Images of mitotic D.Mel cells after depletion of PLP and Ana2 by RNAi. D.Mel cells were depleted of PLP (PLP RNAi), Ana2 (Ana2 RNAi), and were double-depleted with Ana2 and PLP (Ana2 +PLP RNAi) (mCherry RNAi was used as negative control) for three days. Cells were immunostained with anti-SAS-6, Bld10, phospho-H3 and PLP antibodies. Scale bar, 5 μm. (B) Quantification of the SAS-6 intensity per centrosome in the mitotic cells in the indicated RNAi conditions. Means ± s.d are shown in red (***p<0.001, Mann-Whitney U test). Results are representative of three independent experiments (N > 50 centrosomes for each condition). (C) Western blotting analysis of PLP, Ana2 and SAS-6 protein levels in the cells treated with the indicated dsRNAs using the antibodies against PLP, Ana2, SAS-6 and actin (loading control). Red arrowheads indicate expected bands of the longest PLP isoform. (D) Images of mitotic cells, after depletion of PLP and Ana2 by RNAi, used for centriole counting. Cells were immunostained with anti-Bld10 (red), phospho-H3 (red), and SAS-4 (another centriole marker, green) antibodies. Scale bar, 5 μm. (E) Quantification of centriole number per cell (N > 50). Data are the average of three experiments ± s.d (*p<0.05, Mann-Whitney U test performed for the 0–1 centriole category).
-
Figure 6—source data 1
The source data to plot the graph in Figure 6B and E.
- https://doi.org/10.7554/eLife.41418.024
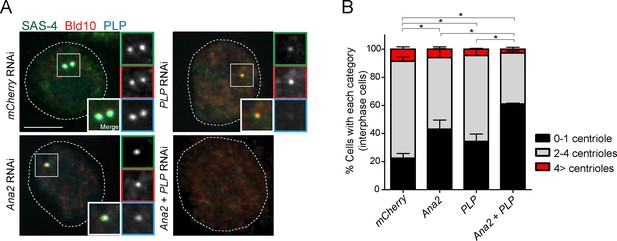
Counting of the centriole number in interphase cells.
(A) Images of interphase D. Mel cells after depletion of PLP and Ana2 by RNAi, used for centriole counting in Figure 6. Cells were immunostained with anti-Bld10 (red), SAS-4 (another centriole marker, green), and PLP (cyan) antibodies. Scale bar, 5 μm. (B) Quantification of centriole number per cell (N > 50). Data are the average of three experiments ± s.d (*p<0.05, Mann-Whitney U test performed for the 0–1 centriole category).
-
Figure 6—figure supplement 1—source data 1
The source data to plot the graph in Figure 6—figure supplement 1B.
- https://doi.org/10.7554/eLife.41418.021
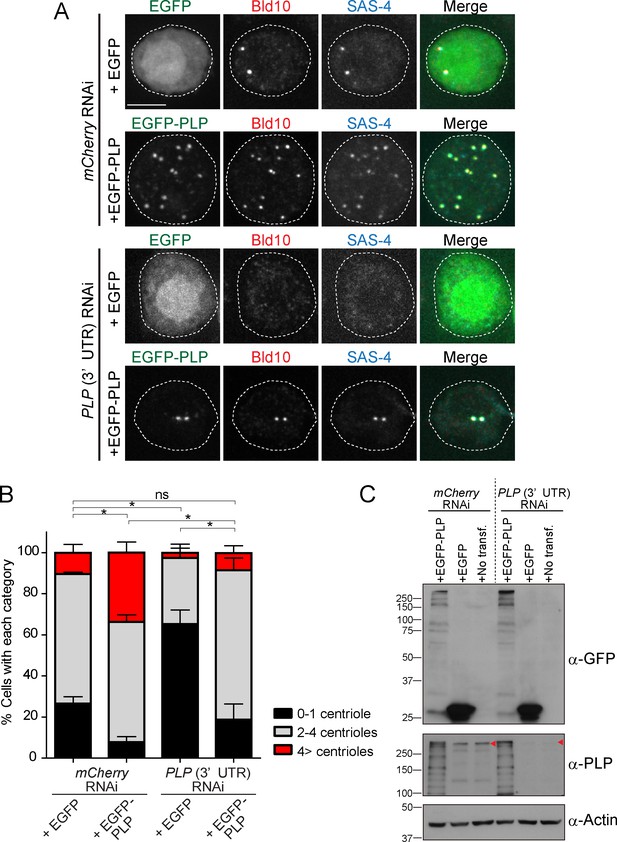
Expression of full-length PLP rescues centriole number defects in PLP-depleted cells.
(A) Images of interphase D.Mel cells expressing EGFP-tagged full-length PLP or only EGFP after depletion of endogenous PLP by RNAi. D.Mel cells were depleted of PLP (PLP 3’UTR RNAi) (mCherry RNAi was used as negative control) for three days for two rounds. Cells were immunostained with anti-Bld10 (red) and SAS-4 (cyan). Scale bar, 5 μm. (B) Quantification of centriole number per cell (N > 50) in EGFP-positive cells shown in A. Data are the average of three experiments ± s.d (ns, not significant, *p<0.05, Mann-Whitney U test performed for the 0–1 centriole category). (C) Western blotting analysis of in the cells shown in A using the antibodies against GFP, PLP and actin (loading control). Red arrowheads indicate expected bands of the longest PLP isoform.
-
Figure 6—figure supplement 2—source data 1
The source data to plot the graph in Figure 6—figure supplement 2B.
- https://doi.org/10.7554/eLife.41418.023
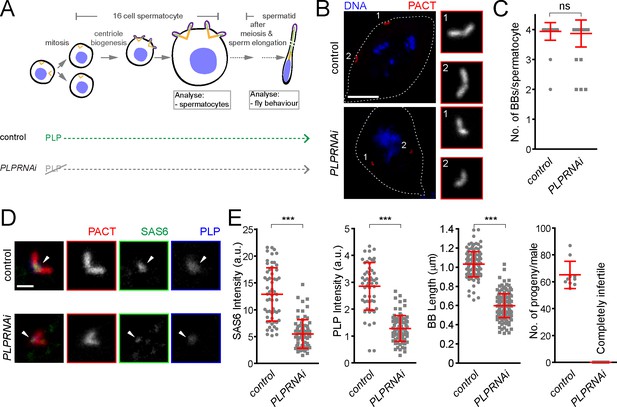
Drosophila pericentrin (PLP) is required for SAS-6 recruitment to the sperm centriole/basal body and for its elongation.
(A) Schematic illustration of the experiments to deplete PLP during centriole biogenesis and elongation. (B) Representative images of mature spermatocytes in flies with different genotypes. PACT (red) is a commonly used marker for basal bodies (BB) and DAPI (blue) stains DNA. Insets show RFP::PACT close to the numbers (in gray scale). (C) Quantification of the number of BBs per cell in mature spermatocytes. (D) Representative images of mature spermatocyte BB in flies with different genotypes. RFP::PACT (red) marks BBs, Anti-SAS-6 (green) and Anti-PLP (blue) antibodies stain the proximal part of BB (arrowheads). (E) Quantification of SAS-6, PLP, BB length and the number of progeny in flies with different genotypes. We repeated all experiments three times. For SAS-6 and PLP intensities and BB length analysis, the number of BBs quantified for each genotype is N ≥ 108 (54 pairs of BBs) and N ≥ 128, respectively. The total number of males used for each histogram bar is N ≥ 10. Notably, given that moderate overexpression of PACT domain (using polyUbiquitin promoter) in the plp mutant fly fails to rescue the observed centriole as well as behavior defects of the mutant (Martinez-Campos et al., 2004), we used RFP::PACT to study the sperm basal bodies in the knockdown experiments. Scale bars in (B) and (D) represent 10 and 1 µm, respectively. Means ± s.d are shown in red (ns-not significant, ***p<0.001, Mann-Whitney U test).
-
Figure 7—source data 1
The source data to plot the graph in Figure 7C and E.
- https://doi.org/10.7554/eLife.41418.026
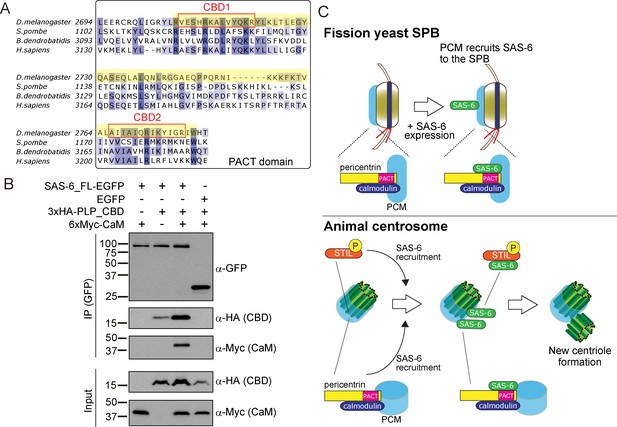
SAS-6 interacts with the calmodulin binding domain within PACT.
(A) Graphical representations of the calmodulin (CaM)-binding domains (CBD1 and CBD2) within the multiple sequence alignment of the PACT domain of pericentrin proteins in the indicated species. CBD1 and CBD2 are marked with red squares. In yellow, minimal fragment containing both CBD1 and CBD2 (hereafter called CBD and used for subsequent experiments). The sequences of the PACT domain were aligned using Clustal Omega multiple sequence alignment tool and visually represented using Jalview software (McWilliam et al., 2013; Waterhouse et al., 2009). The alignments are color-coded in shades of blue for the percentage identity of amino acids; darkest blue (>80%), mid blue (>60%), light blue (>40%), white (<=40%). (B) Complex formation between SAS-6, the highly conserved CBD within PACT domain and calmodulin. Protein extract was prepared from the D.Mel cells expressing SAS6-EGFP or EGFP, HA-tagged CBD and Myc-tagged calmodulin. The GFP-tagged proteins were immunoprecipitated with anti-GFP antibody. Immunoprecipitates and inputs (20%) were analyzed by western blotting using the indicated antibodies. (C) Schematic representation of the ancestral role of the PCM in recruiting centriole proteins, centriole biogenesis and elongation. When SAS-6 was heterologously expressed in fission yeast cells, it localized to the SPB through interaction with Pcp1/pericentrin (upper panel). This revealed a novel interaction between SAS-6 and pericentrin in animals, which is important for centriole biogenesis (lower panel). It is likely that SAS-6 is recruited to the pre-existing centriole prior to new centriole formation by two complementary pathways: PLK4-STIL/Ana2 and pericentrin. Eventhough yeast and animals are separated by one billion years of evolution, the pericentrin/Pcp1-SAS-6 interaction surface has been conserved, likely because the binding of pericentrin to calmodulin constrained its evolution.

Intensity of Sid4 is increased in the pcp1 mutant.
(A-F) Quantification of the intensity of the centriole proteins and Sid4-tdTomato per SPB in the indicated conditions. B, D and F correspond to A, C, and E.Means ± s.d. are shown in red (ns-not significant, * p<0.05, ** p<0.001, *** p<0.0001, Mann-Whitney U test).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4- D18 lys1::Patb2-DmSAS-6-GFP-FLAG-6xHis-ura4 + sid4-tdTomato-natMX6 | This paper | DI 456 | Figure 2A,B,Figure 2 —figure supplement 1A,B, Figure 4—figure supplement 3C,D |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmBld10-GFP-FLAG-6xHis-ura4 + sid4-mRFP-natMX6 | This paper | DI 190 | Figure 2A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-6xHis-FLAG-YFP-DmSAS-4-ura4 + sid4-mRFP-natMX6 | This paper | DI 202 | Figure 2A,B |
| Strain (Schizosaccharomyces pombe) | h- ura4-D18 lys1::Pnmt1-6xHis-FLAG-YFP- DmAna2-ura4 + sid4-tdtomato- natMX6 | This paper | DI 655 | Figure 2A,B |
| Strain (Schizosaccharomyces pombe) | h + ura4-D18 leu1::Pnmt41-6xHis-FLAG-GFP-DmPlk4-ura4 + sid4-tdtomato-natMX6 | This paper | DI 665 | Figure 2A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmSAS-6-GFP-FLAG-6xHis-ura4 + sid4 -tdtomato-natMX6 pcp1-HA-hphMX6 cut7-446 | This paper | DI 492 | Figure 2C, Figure 3 |
| Strain (Schizosaccharomyces pombe) | h + leu1-32 ura4-D18 lys1::Patb2-DmBld10-GFP-FLAG-6xHis-ura4 + sid4-tdtomato-natMX6 pcp1-HA-hphMX6 cut7-446 | This paper | DI 706 | Figure 2C, Figure 3 |
| Strain (Schizosaccharomyces pombe) | h + leu1-32 ura4-D18 lys1::Patb2-6xHis- FLAG-YFP-DmSAS-4-ura4 + sid4-tdtomato-natMX6 pcp1-HA-hphMX6 cut7-446 | This paper | DI 709 | Figure 2C, Figure 3 |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-GFP-FLAG-6xHis-ura4 + sid4-tdtomato-natMX6 pcp1-HA-hphMX6 cut7-446 | This paper | DI 486 | Figure 2C |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 | This paper | DI 7 | Figure 2C, Figure 3, Figure 2—figure supplement 1A,B, Figure 4—figure supplement 2A–C, Figure 4—figure supplement 3A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmSAS-6-GFP-FLAG-6xHis-ura4 + sid4-tdtomato-natMX6 pcp1-14-HA-hphMX6 cut7-446 | This paper | DI 631 | Figure 3 |
| Strain (Schizosaccharomyces pombe) | h + leu1-32 ura4-D18 lys1::Patb2-DmBld10-GFP-FLAG-6xHis-ura4 + sid4-tdtomato-natMX6 pcp1-14-HA-hphMX6 cut7-446 | This paper | DI 710 | Figure 3 |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-6xHis-FLAG-YFP-DmSAS-4-ura4 + sid4-tdtomato-natMX6 pcp1-14-HA-hphMX6 cut7-446 | This paper | DI 719 | Figure 3 |
| Strain (Schizosaccharomyces pombe) | h + leu1-32 ura4-D18 lys1::Patb2-DmSAS-6-GFP-FLAG-6xHis-ura4+ | This paper | DI 105 | Figure 4B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmSAS-6-GFP-FLAG-6xHis-ura4 + sfi1-CFP-natMX6 | This paper | DI 636 | Figure 4D,E |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-GFP-FLAG-6xHis-ura4 + sfi1-CFP-natMX6 | This paper | DI 638 | Figure 4D,E |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmSAS-6- GFP-FLAG-6xHis-ura4 + sid4-tdTomato-natMX6 | This paper | DI 456 | Figure 2—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmBld10- GFP-FLAG-6xHis-ura4 + sid4-mRFP-natMX6 | This paper | DI 190 | Figure 2—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-6xHis-FLAG-YFP-DmSAS-4-ura4 + sid4-mRFP-natMX6 | This paper | DI 202 | Figure 2—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- ura4-D18 lys1::Pnmt1- 6xHis-FLAG -YFP-DmAna2-ura4 + sid4- tdtomato-natMX6 | This paper | DI 655 | Figure 2—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 | This paper | DI 7 | Figure 2—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 pcp1- GFP-kanMX6 sid4-tdtomato-natMX6 | This paper | DI 547 | Figure 4—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 arg1::Pnmt1-DmSAS-6-GFP-FLAG-6xHis-kanMX6 sid4- tdtomato -natMX6 | This paper | DI 646 | Figure 4—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 arg1::Pnmt1-DmSAS-6-GFP-FLAG-6xHis-kanMX6 pcp1-tdTomato-natMX6 | This paper | DI 721 | Figure 4—figure supplement 1A,B |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 arg1::Pnmt1-DmSAS-6-GFP-FLAG-6xHis-kanMX6 sid4-tdtomato-natMX6 | This paper | DI 646 | Figure 4—figure supplement 2A–C |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 arg1::Pnmt1-DmSAS-6(Reg7, 1–176 aa) -GFP-FLAG-6xHis-kanMX6 sid4-tdTomato-natMX6 | This paper | DI 671 | Figure 4—figure supplement 2A–C |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 arg1::Pnmt1-DmSAS-6 (Reg6, 177–472 aa)- GFP-FLAG-6xHis-kanMX6 sid4-tdtomato-natMX6 | This paper | DI 648 | Figure 4—figure supplement 2A–C |
| Strain (Schizosaccharomyces pombe) | h- leu1-32 ura4-D18 lys1::Patb2-DmSAS-6- GFP-FLAG-6xHis-ura4 + sid4-tdTomato-natMX6 pcp1-HA- hphMX6 cdc25-22 | This paper | DI 454 | Figure 4—figure supplement 3A,B |
| Recombinant DNA reagent (plasmid) | pLYS1U-GFH21c-DmSAS-6 | This paper | Expression of DmSAS-6-GFP in S. pombe (atb2 promoter) | |
| Recombinant DNA reagent (plasmid) | pLYS1U-GFH21c-DmBLD10 | This paper | Expression of DmBld10-GFP in S. pombe (atb2 promoter) | |
| Recombinant DNA reagent (plasmid) | pLYS1U-HFY21c-DmSAS-4 | This paper | Expression of YFP-DmSAS-4 in S. pombe (atb2 promoter) | |
| Recombinant DNA reagent (plasmid) | pLYS1U-HFY1c-DmAna2 | This paper | Expression of YFP-DmAna2 in S. pombe (nmt1 promoter) | |
| Recombinant DNA reagent (plasmid) | pDUAL2-HFG1c-DmPlk4 | This paper | Expression of GFP-DmPlk4 in S. pombe (nmt1 promoter) | |
| Recombinant DNA reagent (plasmid) | pARG1-GFH1c-DmSAS-6 | This paper | Expression of DmSAS-6-GFP in S. pombe (nmt1 promoter) | |
| Recombinant DNA reagent (plasmid) | pARG1-GFH1c-DmSAS-6(Reg7, 1–176 aa) | This paper | Expression of DmSAS-6-GFP (Reg7, 1–176 aa) in S. pombe (nmt1 promoter) | |
| Recombinant DNA reagent (plasmid) | pARG1-GFH1c-DmSAS-6(Reg6, 177–472 aa) | This paper | Expression of DmSAS-6-GFP (Reg6, 177–472 aa) in S. pombe (nmt1 promoter) | |
| Recombinant DNA reagent (plasmid) | pREP41-pcp1-mcherry | This paper | Expression of Pcp1-mCherry (full length) in S. pombe (nmt41 promoter) | |
| Recombinant DNA reagent (plasmid) | pREP41-pcp1 (N 1–419)-mcherry | This paper | Expression of Pcp1-mCherry (N 1–419) inS. pombe (nmt41 promoter) | |
| Recombinant DNA reagent (plasmid) | pREP41-pcp1 (M 420–782)-mcherry | This paper | Expression of Pcp1-mCherry (M 420–782) in S. pombe (nmt41 promoter) | |
| Recombinant DNA reagent (plasmid) | pREP41-pcp1 (C 783–1208)-mcherry | This paper | Expression of Pcp1-mCherry (C 783–1208) in S. pombe (nmt41 promoter) | |
| Recombinant DNA reagent (plasmid) | pAWG-DmSAS-6 | This paper | Expression of DmSAS-6-EGFP in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pAGW | This paper | DGRC:1071 | Expression of EGFP in D.Mel cells (actin5c promoter) |
| Recombinant DNA reagent (plasmid) | pAHW-DmPLP_PACT | This paper | Expression of 3xHA-DmPLP_PACT in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pAHW-DmPLP_CBD | This paper | Expression of 3xHA-DmPLP_CBD in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pAMW-DmCaM | This paper | Expression of 6xMyc-DmCaM in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pAGW-DmPLP_PACT | This paper | Expression of EGFP-DmPLP_PACT in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pAGW-DmPLP_FL | This paper | Expression of EGFP-DmPLP_FL in D.Mel cells (actin5c promoter) | |
| Recombinant DNA reagent (plasmid) | pDEST15-DmSAS-6 | This paper | Expression of GST-DmSAS-6 (full length) in E. coli | |
| Recombinant DNA reagent (plasmid) | pGEX6p-1 | GE Healthcare | Expression of GST in E. coli | |
| Recombinant DNA reagent (plasmid) | pET30b-DmPLP_PACT | This paper | Expression of 6xHis-DmPLP_PACT in E. coli | |
| Cell line (Drosophila melanogaster) | D.Mel cells | Thermo Fisher Scientific | ATCC Cat# CRL-1963, RRID:CVCL_Z232 | Drosophila cultured cells |
| Sequence-based reagent | PLP-Forward primer (dsRNA synthesis (PLP)) | This paper | TAATACGACTCACTATAGGGAGAGGAGCGCCTAAAGAACAGTG | |
| Sequence- based reagent | PLP-Reverse primer (dsRNA synthesis (PLP)) | This paper | TAATACGACTCACTATAGGGAGACTGATCGAGCTGTTTGTGGA | |
| Sequence-based reagent | Ana2-Forward primer (dsRNA synthesis (Ana2)) | This paper | GAATTAATACGACTCACTATAGGGAGAATGTTTGTTCCCGAAACGGAGG | |
| Sequence-based reagent | Ana2-Reverse primer (dsRNA synthesis (Ana2)) | This paper | GAATTAATACGACTCACTATAGGGAGACAGAGCCGCCAGATCACTCTTA | |
| Sequence-based reagent | mCherry-Forward primer (dsRNA synthesis (mCherry)) | This paper | ATAATACGACTCACTATAGGGATGGTGAGCAAGGG | |
| Sequence-based reagent | mCherry-Reverse primer (dsRNA synthesis (mCherry)) | This paper | ATAATACGACTCACTATAGGGGTTGACGTTGTAGG | |
| Sequence-based reagent | plp_3UTR_Forward primer (dsRNA synthesis (PLP_3'UTR)) | This paper | TAATACGACTCACTATAGGGAGAGCCCAGGATAGCAGAGTTGAG | |
| Sequence-based reagent | plp_3UTR_Reverse primer (dsRNA synthesis (PLP_3'UTR)) | This paper | TAATACGACTCACTATAGGGAGACGAATGTGAAATAAATTTGGTTTAA | |
| Strain (Drosophila melanogaster) | w1118; Ubq-RFP::PACT;+ | Carvalho-Santos et al., 2010 | ||
| Strain (Drosophila melanogaster) | w1118; +; bamGal4 | Chen and McKearin, 2003 | ||
| Strain (Drosophila melanogaster) | yv; +; UAS-mCherryRNAi | Perkins et al., 2015 | ||
| Strain (Drosophila melanogaster) | w1118; UAS-PLPRNAi; + | Dietzl et al., 2007 | ||
| Strain (Drosophila melanogaster) | w1118; Ubq-RFP::PACT/+; UAS-mCherryRNAi/ bamGal4 | This paper | ||
| Strain (Drosophila melanogaster) | w1118; Ubq-RFP::PACT/UAS-PLPRNAi; bamGal4/+ | This paper | ||
| Antibody | anti-GFP (rabbit polyclonal) | Abcam | Abcam Cat# ab290, RRID:AB_303395 | WB 1:1000 |
| Antibody | anti-RFP (rat monoclonal) | Chromotek | RRID:AB_2336064 | WB 1:1000 |
| Antibody | anti-HA (rat monoclonal) | Roche | Roche Cat# 11867431001, RRID:AB_390919 | WB 1:1000 |
| Antibody | anti-Cdc2 PSTAIRE (rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-53, RRID:AB_2074908 | WB 1:2000 |
| Antibody | anti-Drosophila SAS-6 (rabbit polyclonal) | Gift from J Gopalakrishnan | WB 1:500 | |
| Antibody | anti-Drosophila SAS-6 (rat polyclonal) | Gift from N Dzhindzhev and D Glover | IF 1:500 | |
| Antibody | anti-Drosophila Ana2 (rat polyclonal) | Gift from N Dzhindzhev and D Glover | WB 1:4000 | |
| Antibody | anti-phospho Histone H3 (Ser10) (rabbit polyclonal) | Millipore | Millipore Cat# 06–570, RRID:AB_310177 | IF 1:2000 |
| Antibody | anti-Drosophila Bld10 (rabbit polyclonal) | Gift from T Megraw | IF 1:5000 | |
| Antibody | anti-Drosophila PLP (guinea pig polyclonal) | Gift from G Rogers | WB 1:1000 | |
| Antibody | anti-Drosophila PLP (chicken polyclonal) | Bettencourt-Dias et al., 2005 | IF 1:500 | |
| Antibody | anti-Actin (rabbit polyclonal) | Sigma-Aldrich | Sigma-Aldrich Cat# A2066, RRID:AB_476693 | WB 1:2000 |
| Antibody | anti-GST (mouse monoclonal) | Cell Signaling Technology | Cell Signaling Technology Cat# 3513, RRID:AB_1642209 | WB 1:1000 |
| Antibody | anti-His-tag (mouse monoclonal) | Millipore | Millipore Cat# 70796–3, RRID:AB_11213479 | WB 1:1000 |
| Antibody | anti-Myc (9E10) (mouse monoclonal) | Santa Cruz Bio technology | Santa Cruz Biotechnology Cat# sc-40, RRID:AB_627268 | WB 1:1000 |
| Antibody | anti-Rat IgG (secondary, DyLight 488, Donkey) | Bethyl Laboratories | IF 1:100 | |
| Antibody | anti-Rabbit IgG (secondary, Rhodamine-Red, Donkey) | Jackson ImmunoResearch | IF 1:100 | |
| Antibody | anti-Rabbit IgG (secondary, Cy5, Donkey) | Jackson ImmunoResearch | IF 1:100 | |
| Antibody | anti-Chicken IgY (secondary, Cy5, Donkey) | Jackson ImmunoResearch | IF 1:100 | |
| Antibody | anti-Rat IgG (secondary, Cy5, Donkey) | Jackson ImmunoResearch | IF 1:100 | |
| Antibody | anti-Mouse IgG (secondary, HRP-conjugated, Donkey) | Jackson ImmunoResearch | WB 1:5000 | |
| Antibody | anti-Rabbit IgG (secondary, HRP-conjugated, Donkey) | Jackson ImmunoResearch | WB 1:5000 | |
| Antibody | anti-Guinea pig IgG (secondary, HRP-conjugated, Donkey) | Jackson ImmunoResearch | WB 1:5000 | |
| antibody | anti-Rat IgG (secondary, HRP-conjugated, Goat) | Bethyl Laboratories | WB 1:5000 | |
| antibody | anti-Rat IgG (secondary, IRDye 800CW, Goat) | LI-COR | WB 1:10000 | |
| antibody | anti-Mouse IgG (secondary, IRDye 680CW, Goat) | LI-COR | WB 1:10000 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41418.028





