Translesion polymerase kappa-dependent DNA synthesis underlies replication fork recovery
Figures
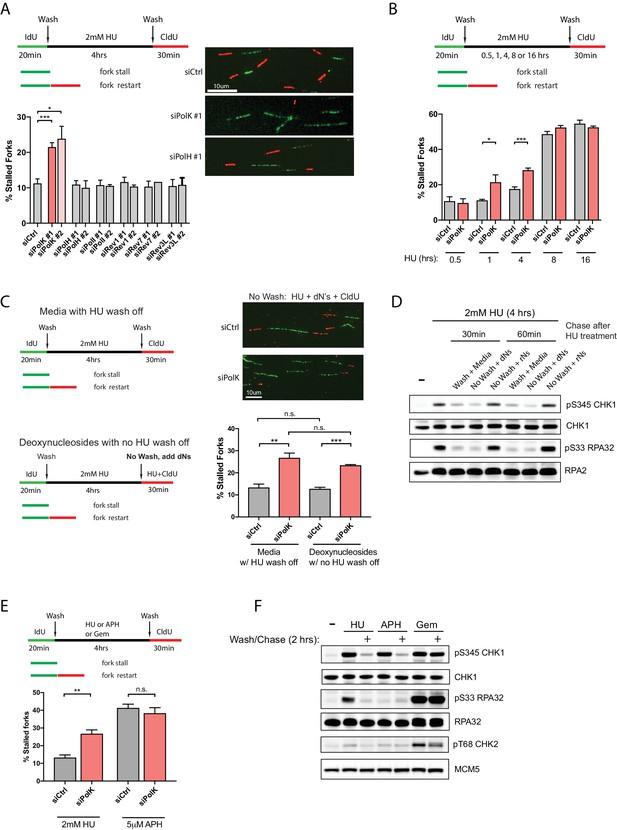
PolK is required for replication fork restart due to nucleotide deprivation.
(A) Schematic for measuring replication fork restart by DNA fiber analysis. Quantification of fork restart efficiency (% stalled forks) in HU-treated (2 mM) RPE-1 cells using two independent siRNAs against individual TLS Pols as indicated. Representative images of the DNA fiber tracts are shown. (B) Quantification of fork restart efficiency in RPE-1 cells comparing different HU (2 mM) treatment time-points in the presence or absence of PolK siRNA knockdown. (C) Quantification of fork restart efficiency in HU-treated RPE-1 cells with either a wash step with fresh media or with no wash (HU still present) supplemented with 250 μM deoxynucleosides (dNs) for recovery. (D) Western blot analysis of RPE-1 cells treated with 2 mM HU for 4 hr followed by either a wash step with fresh media or no wash (HU still present) supplemented with 250 μM deoxynucleosides (dNs) or 250 μM ribonucleosides (rNs) for 30 or 60 min chase. (E) Quantification of fork restart efficiency comparing fork-stalling agents, HU (2 mM) or APH (5 μM), in the presence or absence of PolK siRNA knockdown. (F) Western blot analysis of RPE-1 cells treated with either HU (2 mM), APH (5 μM), or Gemcitabine (Gem, 1 μM) for 4 hr, followed by a wash step and recovery in fresh media for 2 hr. Data for % stalled forks are represented by mean ± s.d. of three independent experiments and p-values calculated using t-test with Welch’s correction. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
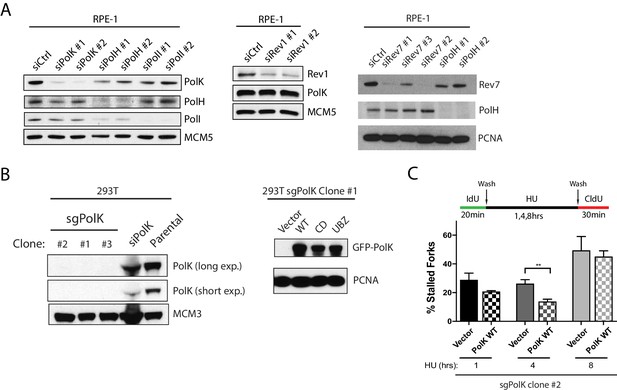
siRNA knockdown efficiencies and complementation of CRISPR 293T sgPolK clonal cells.
https://doi.org/10.7554/eLife.41426.003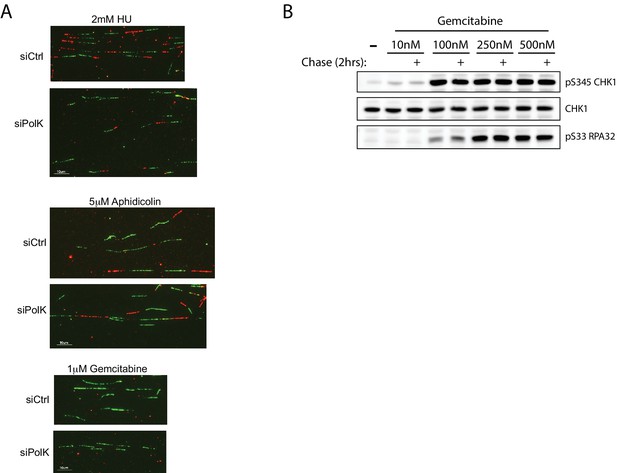
Gemcitabine-induced stalled forks are not amenable for fork restart assays.
https://doi.org/10.7554/eLife.41426.004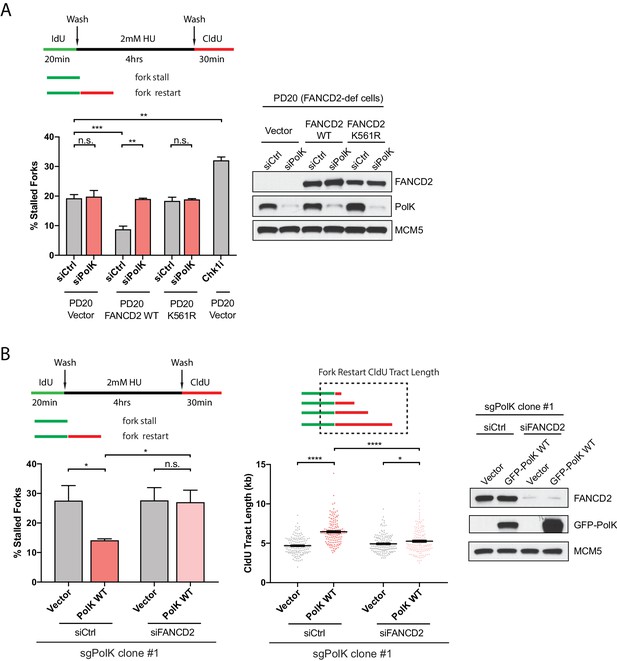
PolK functions in concert with the FA pathway to promote replication fork restart.
(A) Quantification of fork restart efficiency in FANCD2-deficient patient cells (PD20) complemented with either vector only, FANCD2 WT, or K561R mutant in the presence or absence of PolK siRNA and treated as indicated. PD20 (vector only) cells were treated with 300 nM Chk1i (AZD7762) throughout the duration of HU and CldU time points as a positive control for the detection of elevated fork-stalling events. Western blot analysis showing siRNA knockdown efficiency in PD20 cells. (B) Quantification of fork restart efficiency in 293T CRISPR PolK (sgPolK) cells complemented with either empty vector or GFP-PolK WT in the presence or absence of FANCD2 siRNA and treated as indicated. CldU (red) tract length measurements of restarted forks determine the varying degree of individual fork restart events. Western blot analysis showing expression and siRNA knockdown efficiency in sgPolK 293 T cells. Data for % stalled forks are represented by mean ± s.d. of three independent experiments and p-values calculated using t-test with Welch’s correction. Data for tract length measurements are plotted from three independent experiments with mean ± s.e.m. and p-values calculated using Mann-Whitney t-test. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
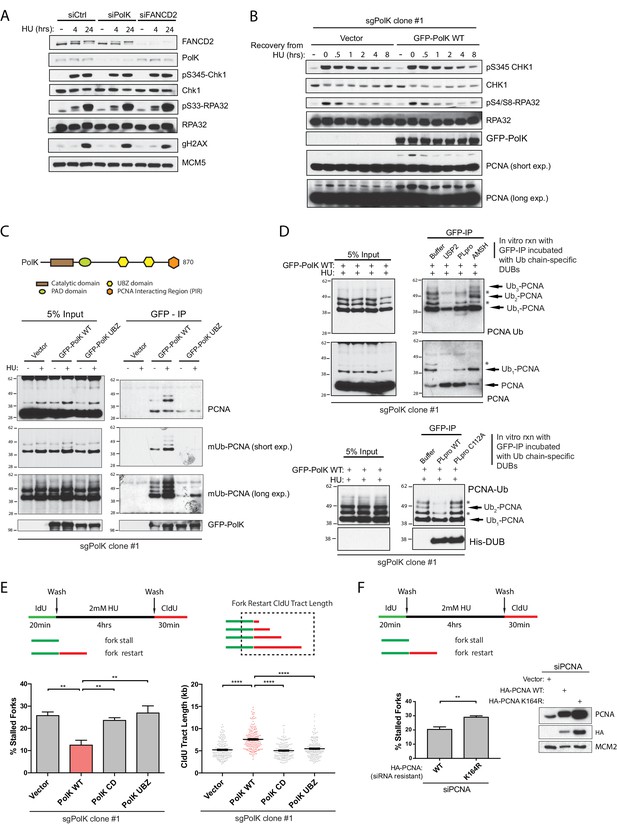
PolK interacts with K48-linked polyubiquitinated PCNA via its UBZ domains in a HU-dependent manner.
(A) Western blot analysis of RPE-1 cells treated with the indicated siRNAs and HU (2 mM) time-points. (B) Western blot analysis of 293T sgPolK cells complemented with either empty vector or GFP-PolK WT and pulsed with HU (2 mM) for 4 hr before wash step and recovery for the indicated time-points. (C) Schematic diagram showing domains of PolK. Formaldehyde-induced crosslinking of 293T sgPolK cells treated with HU (2 mM) for 4 hr as indicated. Extracts from cells complemented with either empty vector, GFP-PolK WT or a double ubiquitin-binding domain mutant (UBZ) of GFP-PolK were then subjected to anti-GFP pulldown, followed by Western blot analysis with the indicated antibodies. (D) Ubiquitin chain restriction digest analysis using similarly treated and immunoprecipitated (IP) samples as in (C) to enrich for polyubiquitinated PCNA that is bound by GFP-PolK and induced by HU. Samples on beads were then incubated with 900 ng of indicated recombinant DUBs for 1 hr at 37°C prior to Western blot analysis with the indicated antibodies (upper and lower panels). SARS PLpro catalytic mutant (C112A) was used for negative control as indicated (lower panel). (E) Quantification of fork restart efficiency in 293T sgPolK cells complemented with either empty vector, GFP-PolK WT, Catalytic-Dead (CD), or ubiquitin-binding mutant (UBZ). CldU (red) tract length measurements of restarted forks were determined for WT and the different PolK mutants. (F) Quantification of fork restart efficiency in U2OS cells treated with PCNA siRNA and complemented with siRNA-resistant HA-tagged PCNA-WT or ubiquitin site mutant HA-PCNA K164R. Western blot analysis showing exogenously expressed siRNA-resistant HA-PCNA in U2OS cells. Data for % stalled forks are represented by mean ± s.d. of three independent experiments and p-values calculated using t-test with Welch’s correction. Data for tract measurements are plotted from three independent experiments with mean ± s.e.m. and p-values calculated using Mann-Whitney t-test. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
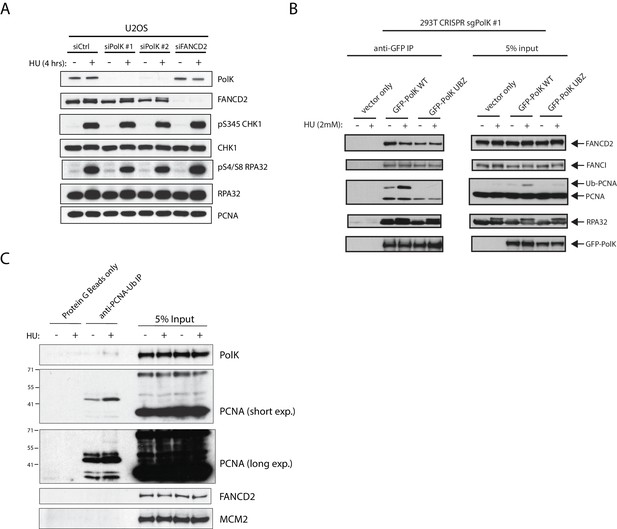
PolK interacts with FANCD2 and RPA independently of its UBZ domain.
https://doi.org/10.7554/eLife.41426.007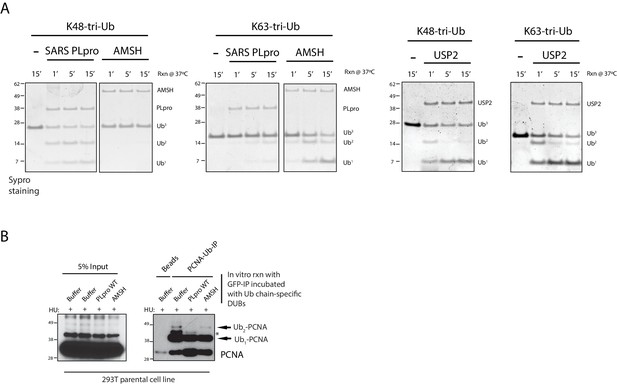
HU-dependent PCNA polyubiquitination is susceptible to an in vitro K48-specific polyUb DUB cleavage reaction.
https://doi.org/10.7554/eLife.41426.008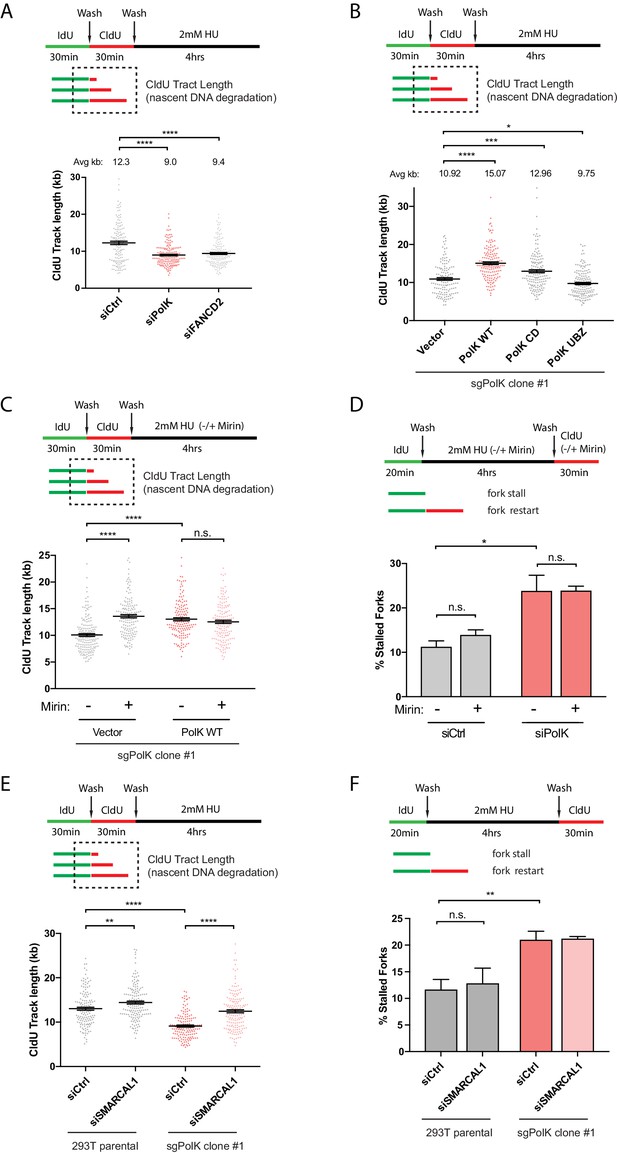
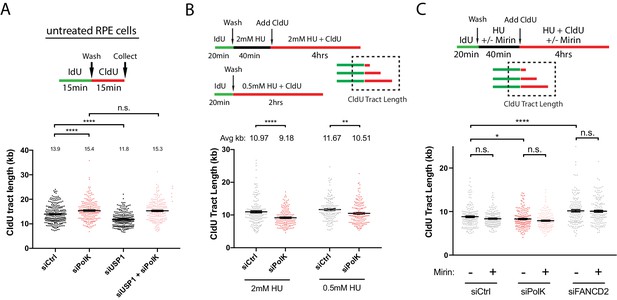
PolK prevents MRE11-dependent nascent DNA degradation.
(A) Schematic for measuring nascent DNA degradation (shortened CldU-labeled tracts) by DNA fiber analysis (A–C,E). Quantification of nascent DNA degradation (changes in CldU tract lengths) in 293 T cells treated with the indicated siRNAs. (B) Quantification of nascent DNA degradation in 293T sgPolK cells that were complemented with either empty vector, GFP-PolK WT or the indicated GFP-PolK mutants. (C) Quantification of nascent DNA degradation in 293T sgPolK cells complemented with either empty vector or GFP-PolK WT were treated with or without Mre11 inhibitor, Mirin (50 μm), in the presence of HU as indicated. (D) Quantification of fork restart efficiency in RPE-1 cells with the indicated siRNAs and treated with or without Mirin (50 μM) in the presence of HU (2 mM) as indicated. (E) Quantification of nascent DNA degradation in parental 293T or 293T sgPolK cells treated with the indicated siRNAs. (F) Quantification of fork restart efficiency in parental 293T or 293T sgPolK cells treated with the indicated siRNAs. Data for % stalled forks are represented by mean ± s.d. of three independent experiments and p-values calculated using t-test with Welch’s correction. Data for tract length measurements are plotted from three independent experiments with mean ± s.e.m. and p-values calculated using Mann-Whitney t-test. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
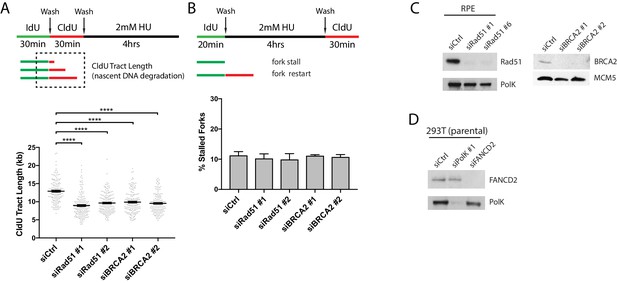
RAD51 and BRCA2 depletion in RPE-1 cells has no effect on replication fork restart after HU treatment.
https://doi.org/10.7554/eLife.41426.010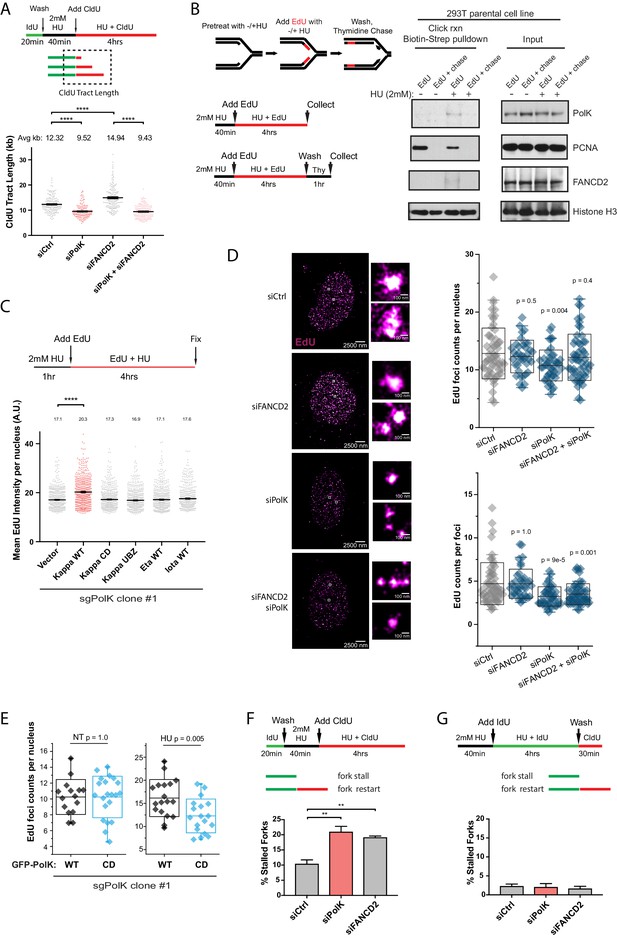
PolK-dependent DNA synthesis in the presence of HU.
(A) Schematic for measuring replication fork speed (CldU tract length) under HU treatment for 4 hr. To ensure that the CldU-labeled DNA was under constant high dose HU treatment, cells were pre-treated with 2 mM HU for 40 min prior to the addition of CldU in the presence of HU for 4 hr. Quantification of fork speed in RPE-1 cells treated with the indicated siRNAs. (B) Schematic of a modified iPOND assay to measure proteins associated with the replisome under nucleotide starvation conditions. Similar to (A), 293 T cells were either left untreated or pretreated with HU (2 mM) for 40 min prior to the addition of EdU (10 μM) in the presence of HU for 4 hr. Cells were then either collected immediately (EdU samples) or chased with Thymidine (10 μM) for 1 hr (EdU + chase) in the presence or absence of HU. Samples without HU were treated for only 10 min with EdU or chased with Thymidine for 1 hr. Western blot analysis showing the biotin-streptavidin pulldown after click-reaction in parental 293 T cells and probed with the indicated antibodies. (C) Schematic for measuring EdU incorporation intensity by direct fluorescence measurements in HU-treated cells. U2OS sgPolK cells were complemented with either empty vector, GFP-PolK WT, GFP-PolK mutant constructs, or different Y-family TLS Pols, GFP-Pol eta or GFP-Pol iota. Cells were pretreated with HU (2 mM) for 1 hr, prior to the addition of EdU (10 μM) in the presence of HU for 4 hr. Mean EdU intensity per nucleus measured by ImageJ were plotted from three independent experiments. (D) Single-molecule localization imaging of EdU signal distribution per foci or nuclei. RPE-1 cells were treated with the indicated siRNAs prior to pretreatment with HU (2 mM) for 1 hr, followed by the addition of EdU in the presence of HU for 4 hr. Representative super-resolution images of nuclei with EdU signal in magenta are shown. Quantification of EdU foci counts per nuclei and amount of EdU counts per foci are plotted from three independent experiments. (E) U2OS sgPolK cells complemented with either GFP-PolK WT or GFP-PolK CD were pulse-labeled with EdU and treated with HU (2 mM) or not (NT). Treatment conditions and quantification of EdU foci per nuclei by super-resolution imaging techniques were done as in (D). (F) Quantification of fork restart efficiency in RPE-1 cells treated with the indicated siRNAs whereby ‘restarted’ forks are measured as previously elongating forks (IdU tracts) that become converted to CldU tracts in the presence of HU (2 mM). Cells were pretreated with HU for 40 min prior to the addition of CldU for 4 hr to ensure that CldU pulse-labeled cells were already under constant high-dose HU treatment. (G) Quantification of fork restart efficiency in RPE-1 cells treated with the indicated siRNAs whereby IdU pulse-labeled forks under constant HU treatment are measured to determine whether they can be ‘restarted’ after HU wash off (CldU pulse-label). Data for % stalled forks and quantification of EdU foci counts by single-molecule localization imaging are represented by mean ± s.d. of three independent experiments and p-values calculated using t-test with Welch’s correction and indicated above the plots. Data for tract length measurements are plotted from three independent experiments with mean ± s.e.m. and p-values calculated using Mann-Whitney t-test. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
-
Figure 5—source data 1
EdU intensity per nucleus.
- https://doi.org/10.7554/eLife.41426.015
Mirin treatment does not affect replication fork speed in either PolK- or FANCD2-depleted RPE-1 cells.
https://doi.org/10.7554/eLife.41426.012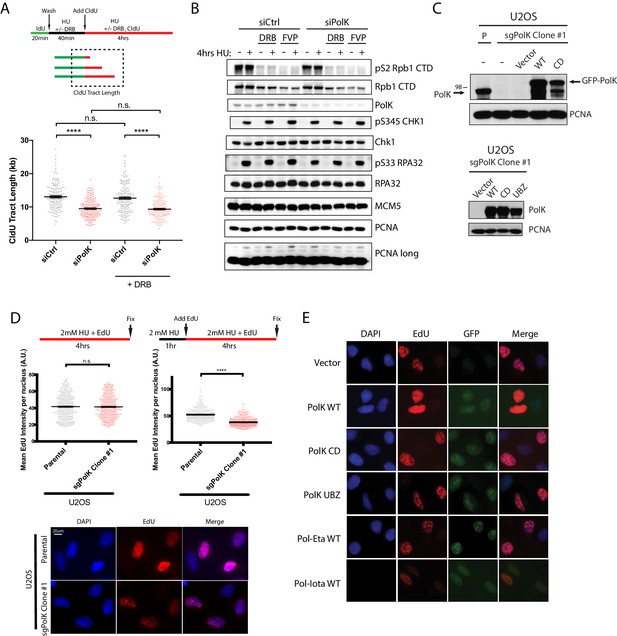
PolK-dependent DNA synthesis under HU is unaffected by DRB treatment.
https://doi.org/10.7554/eLife.41426.013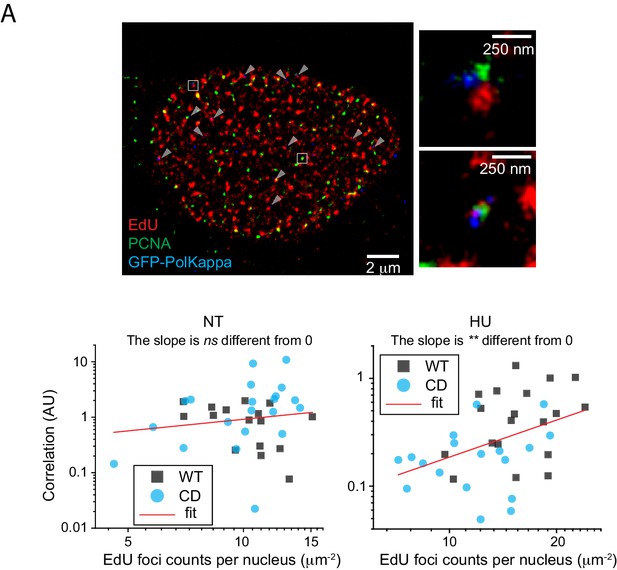
Single-molecule localization image of EdU signal, PCNA, and GFP-PolK in U2OS cells.
https://doi.org/10.7554/eLife.41426.014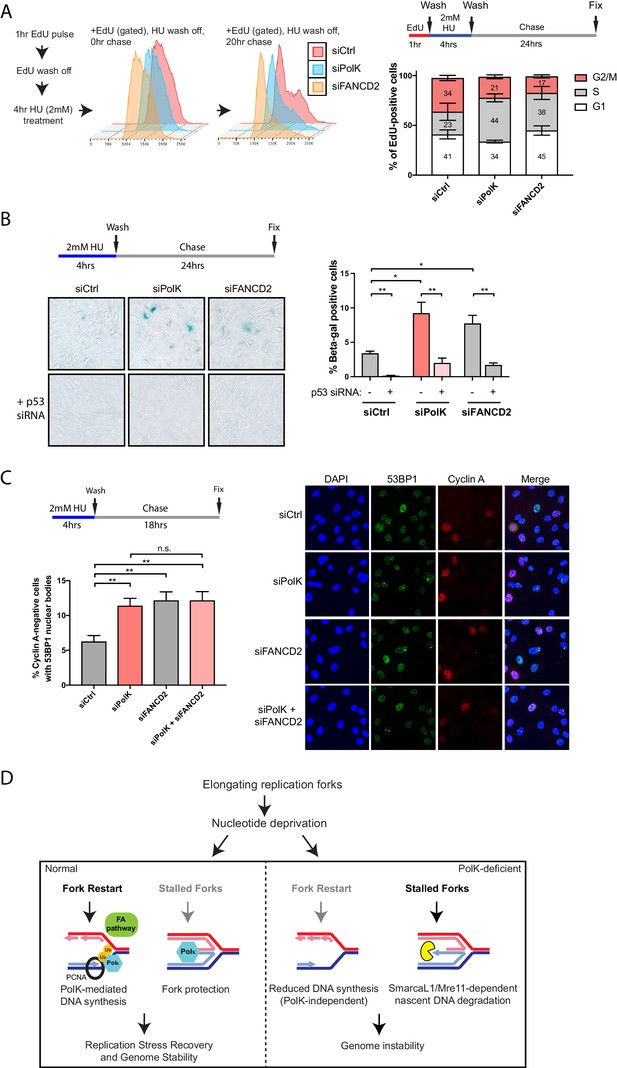
Loss of PolK leads to a p53-dependent cell cycle delay and 53BP1 nuclear body accumulation following HU pulse treatment.
(A) Schematic for measuring cell cycle progression after recovery from HU pulse treatment (A–C). RPE-1 cells treated with the indicated siRNAs were initially pulsed-labeled with EdU (10 μM) for 1 hr to label untreated S-phase cells, followed by a wash step, an HU (2 mM) pulse treatment for 4 hr, another wash step, and recovery (chase) with fresh media for the indicated time. Recovery of EdU-positive, HU pulse-treated cells were tracked by FACS analysis and the proportion of cells in different cell cycle phases were determined by DAPI DNA content (FlowJo). Data represented from three independent experiments with mean ± s.d (A). (B) RPE-1 cells treated with the indicated siRNAs were treated with HU (4 hr), followed by a wash step, and chase for 24 hr with fresh media. Cells were then fixed and stained for SA-β-Gal activity. Data represented from three independent experiments with mean ± s.d., p-value calculated using t-test with Welch’s correction. (C) RPE-1 cells treated with the indicated siRNAs were treated with HU (4 hr), followed by a wash step, and chase for 18 hr with fresh media. Cells were then fixed and co-stained for Cyclin A and 53BP1. Only Cyclin A-negative RPE-1 cells (G1 phase) were quantified for 53BP1 nuclear bodies. Data represented from three independent experiments with a minimum of 300 Cyclin A-negative cells per experiment; mean ± s.d. was plotted and p-value calculated using t-test with Welch’s correction. (D) A model depicting how PolK promotes replication stress recovery and genome stability in an FA pathway-dependent manner in response to conditions of nucleotide starvation. n.s. = no significance, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
-
Figure 6—source data 1
Source data for Figure 6C and B, Figure 6—figure supplement 1B and C.
- https://doi.org/10.7554/eLife.41426.018
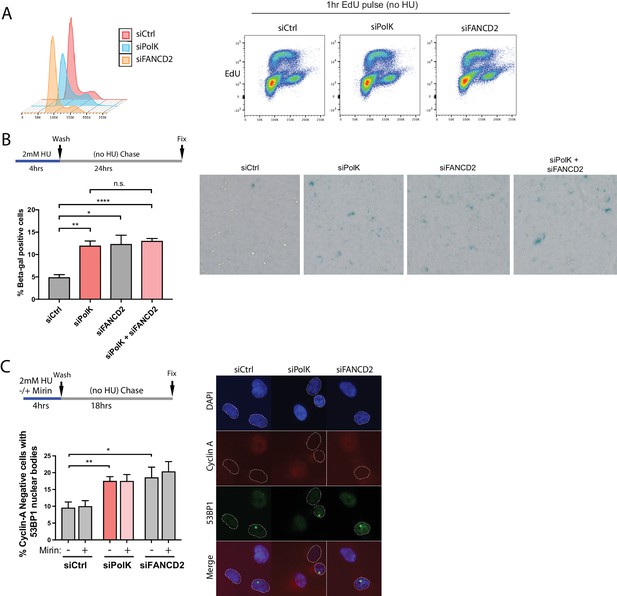
Mirin treatment does not rescue genome instability in HU pulse-treated PolK- or FANCD2-deficient cells.
https://doi.org/10.7554/eLife.41426.017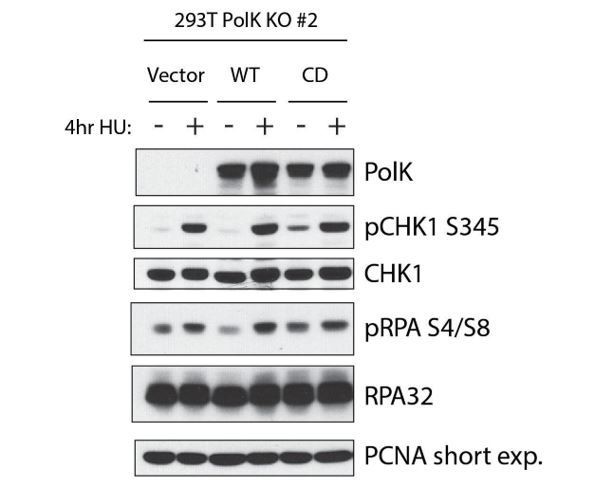
Showing expression of WT vs Catalytic-dead (CD) Pol kappa in Pol kappa sgRNA KO cells.
Even in the absence of HU (2mM), the CD mutant has elevated Chk1 and RPA32 phosphorylation. This is in contrast to the Bétous et al. (2013) study showing that Pol kappa is required for Chk1 activation upon HU treatment.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) female | hTERT RPE-1 (RPE) | ATCC | CRL-4000 | |
| Cell line (Homo sapiens) female | U-2 OS (U2OS) | ATCC | HTB-96 | |
| Cell line (Homo sapiens) female | U2OS PolK KO (U2OS sgPolK Clone #1) | This Paper | CRISPR-Cas9 generated using pX330 construct and guide RNAs listed in Materials and methods. | |
| Cell line (Homo sapiens) | 293T | ATCC | CRL-3216 | |
| Cell line (Homo sapiens) | 293T PolK KO #1 (sgPolK Clone #1) | This Paper | CRISPR-Cas9 generated using pX330 construct and guide RNAs listed in Materials and methods. | |
| Cell line (Homo sapiens) | 293T PolK KO #2 (sgPolK Clone #2) | This Paper | CRISPR-Cas9 generated using pX330 construct and guide RNAs listed in Materials and methods. | |
| Cell line (Homo sapiens) | PD20 Vector | PMID: 11239454 | Garcia-Higuera et al., 2001 | |
| Cell line (Homo sapiens) | PD20 FANCD2 WT | PMID: 11239454 | Garcia-Higuera et al., 2001 | |
| Cell line (Homo sapiens) | PD20 FANCD2 K561R | PMID: 11239454 | Garcia-Higuera et al., 2001 | |
| Transfected construct (Homo sapiens) | pCDNA3.1 HA- PCNA WT | PMID: 22157819 | Dungrawala and Cortez, 2015 | |
| Transfected construct (Homo sapiens) | pCDNA3.1 HA- PCNA K164R | PMID: 22157819 | Dungrawala and Cortez, 2015 | |
| Recombinant DNA reagent | eGFP-C1 | ClonTech | ||
| Transfected construct (Homo sapiens) | eGFP-C1-PolK WT | PMID: 22157819 | Dungrawala and Cortez, 2015 | |
| Transfected construct (Homo sapiens) | eGFP-C1-PolK CD | This Paper | Site directed mutagenesis using primers listed in Materials and methods. D198A/E199A | |
| Transfected construct (Homo sapiens) | eGFP-C1-PolK UBZ | This Paper | Site directed mutagenesis using primers listed in Materials and methods. D644A/D799A | |
| Transfected construct (Homo sapiens) | eGFP-C1-PolH WT | Dungrawala and Cortez, 2015 Dungrawala and Cortez, 2015 PMID: 22157819 | ||
| Transfected construct (Homo sapiens) | eGFP-C1-PolI WT | Dungrawala et al., 2015 PMID: 22157819 | ||
| Recombinant DNA reagent | pX330 | Addgene | #42230 | |
| Antibody | Mouse anti-IdU | BD Bioscience | Cat. #: 347580 | IF (1:200) |
| Antibody | Rat anti-CldU | Abcam | Cat. #: ab6326 | IF (1:100) |
| Antibody | Mouse anti-PCNA | Abcam | Cat. #: ab29 | WB (1:5000) |
| Antibody | Rabbit anti-PCNA-Ub | Cell Signaling | Cat. #: D5C7P | WB (1:1000) |
| Antibody | Mouse anti- FANCD2 | Santa Cruz | Cat. #: sc-20022 | WB (1:500) |
| Antibody | Rabbit anti- FANCD2 | Novus Biological | Cat. #: NB100-182 | WB (1:5000) |
| Antibody | Rabbit anti-pCHK1 S345 | Cell Signaling | Cat. #: 133D3 | WB (1:5000) |
| Antibody | Goat anti-CHK1 | Abcam | Cat. #: ab2845 | WB (1:5000) |
| Antibody | Mouse anti-gH2AX | EMD Millipore | Cat. #: 05–636 | WB (1:5000) |
| Antibody | Mouse anti-GFP | Santa Cruz | Cat. #: sc-9996 | IF (1:200), WB (1:5000) |
| Antibody | Mouse anti-Cyclin A2 | CalBiochem | clone E23 | IF (1:100) |
| Antibody | Rabbit anti-53BP1 | Abcam | Cat. #: ab21083 | IF (1:200) |
| Antibody | Mouse anti-POLK | Santa Cruz | Cat. #: sc-166667 | WB (1:1000) |
| Antibody | Mouse anti-POLH | Santa Cruz | Cat. #: sc-17770 | WB (1:1000) |
| Antibody | Rabbit anti-POLI | Bethyl | Cat. #: A301-304A | WB (1:5000) |
| Antibody | Rabbit anti-MCM2 | Bethyl | Cat. #: A300-094A | WB (1:5000) |
| Antibody | Rabbit anti-MCM5 | Bethyl | Cat. #: A300-195A | WB (1:5000) |
| Antibody | Rabbit anti- Histone H3 | Abcam | Cat. #: ab1791 | WB (1:5000) |
| Antibody | Rabbit anti-REV7 | Abcam | Cat. #: ab180579 | WB (1:5000) |
| Antibody | Rabbit anti-pRPA32 S33 | Bethyl | Cat. #: A300-246A | WB (1:5000) |
| Antibody | Rabbit anti-pRPA32 S4/S8 | Bethyl | Cat. #: A700-009 | WB (1:5000) |
| Antibody | Rabbit anti-RPA32 | Bethyl | Cat. #: A300-244A | WB (1:5000) |
| Antibody | Mouse anti-BRCA2 | CalBiochem | clone 2B | WB (1:1000) |
| Antibody | Mouse anti-HA | BioLegend | Cat. #: 901502 | WB (1:5000) |
| Antibody | Goat anti-HisTag | Bethyl | Cat. #: A190-113A | WB (1:5000) |
| Antibody | Rabbit anti-REV1 | Santa Cruz | Cat. #: sc-48806 | WB (1:1000) |
| Antibody | Rabbit anti-FANCI | Bethyl | Cat. #: A301-254A | WB (1:5000) |
| Antibody | Rabbit anti-MCM4 | Bethyl | Cat. #: A300-193A | WB (1:5000) |
| Antibody | Rabbit anti-MCM3 | Bethyl | Cat. #: A300-192A | WB (1:5000) |
| Antibody | Rabbit anti-Rad51 | Abcam | Cat. #: ab63801 | WB (1:5000) |
| Antibody | Rabbit anti-pRPB1 CTD S2 | Cell Signaling | Cat. #: E1Z3G | WB (1:1000) |
| Antibody | Mouse anti-RPB1 CTD | Cell Signaling | Cat. #: 2629 | WB (1:1000) |
| Antibody | Rabbit anti-pCHK2 T68 | Cell Signaling | Cat. #: C13C1 | WB (1:5000) |
| Recombinant DNA reagent | Fugene 6 Transfection reagent | Promega | E2692 | |
| Recombinant DNA reagent | Lipofectamine RNAiMAX | Invitrogen | Cat #: 13778150 | |
| Sequence- based reagent | CRISPR guide RNAs (sgPolK#1,#2) | This Paper | See Materials and methods | |
| Sequence- based reagent | siRNAs | This Paper | See Materials and methods | |
| Sequence- based reagent | Mutagenesis primers | This Paper | See Materials and methods | |
| Peptide, recombinant protein | Tri-Ubiquitin chains (K48-linked) | Boston Biochem | UC-215B | |
| Peptide, recombinant protein | Tri-Ubiquitin chains (K63-linked) | Boston Biochem | UC-315B | |
| Peptide, recombinant protein | USP2 Catalytic Domain (CD) | Boston Biochem | E-504 | |
| Peptide, recombinant protein | AMSH | Boston Biochem | E-548B | |
| Peptide, recombinant protein | SARS PLPro | Boston Biochem | E-610 | |
| Commercial assay or kit | Click-it EdU Imaging Kit | Invitrogen | C10339 | |
| Commercial assay or kit | Click-it EdU Flow Cytometry Assay Kit | Invitrogen | C10646 | |
| Commercial assay or kit | QuikChange XL Mutagenesis Kit | Agilent | Cat #: 200517 | |
| Commercial assay or kit | TOPO-TA Cloning Kit for Sequencing | Invitrogen | Cat #: 450071 | |
| Chemical compound, drug | Hydroxyurea (HU) | Sigma | H8627 | |
| Chemical compound, drug | Aphidicolin (APH) | Sigma | A4487 | |
| Chemical compound, drug | Gemcitabine (Gem) | Sigma | G6423 | |
| Chemical compound, drug | Mirin | Sigma | M9948 | |
| Chemical compound, drug | AZD7762 (CHK1 inhibitor) | Sigma | SML0350 | |
| Chemical compound, drug | 1-b-D-ribofuranoside (DRB) | Sigma | D1916 | |
| Chemical compound, drug | Flavopiridol (FVP) | Sigma | F3055 | |
| Chemical compound, drug | Formaldehyde 37% w/v | VWR | M134 | |
| Chemical compound, drug | Glycine | Fischer Scientific | BP381 | |
| Chemical compound, drug | 5'-Iodo-2'-deoxyuridine (IdU) | Sigma | I7125 | |
| Chemical compound, drug | 5'-chloro-2'-deoxyuridine (CldU) | Sigma | C6891 | |
| Chemical compound, drug | 5'-ethynyl-2-deoxyuridine (EdU) | Sigma | Cat #: 900584 | |
| Chemical compound, drug | Thymidine (dT) | Sigma | T1895 | |
| Chemical compound, drug | 2'-deoxycytidine HCl (dC) | Sigma | D0776 | |
| Chemical compound, drug | 2'-deoxyadenosine (dA) | Sigma | D8668 | |
| Chemical compound, drug | 2'-deoxyguanosine (dG) | Sigma | D0901 | |
| Chemical compound, drug | Cytidine (rC) | Sigma | C4654 | |
| Chemical compound, drug | Adenosine (rA) | Sigma | A4036 | |
| Chemical compound, drug | Guanosine (rG) | Sigma | G6264 | |
| Chemical compound, drug | Uridine (rU) | Sigma | U3003 | |
| Chemical compound, drug | Anti-GFP mAb agarose beads | MBL | Cat #: D153-8 | |
| Chemical compound, drug | Dynabeads Myone Streptavidin T1 | ThermoFisher Scientific | Cat #: 65601 | |
| Chemical compound, drug | Biotin Azide | ThermoFisher Scientific | Cat #: B10184 | |
| Chemical compound, drug | Protein G beads agarose | ThermoFisher Scientific | Cat #: 20399 | |
| Chemical compound, drug | SYPRO Ruby Protein Gel Stain | ThermoFisher Scientific | Cat #: S12000 | |
| Chemical compound, drug | cOmplete Mini Protease Inhibitor Cocktail | Sigma | Cat #: 11836170001 (Roche) | |
| Software, algorithm | GraphPad Prism | (https://graphpad.com) | RRID:SCR_015807 | |
| Software, algorithm | ImageJ | (https://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | FlowJo | (https://www.flowjo.com/) | RRID:SCR_008520 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41426.019





