Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling
Figures
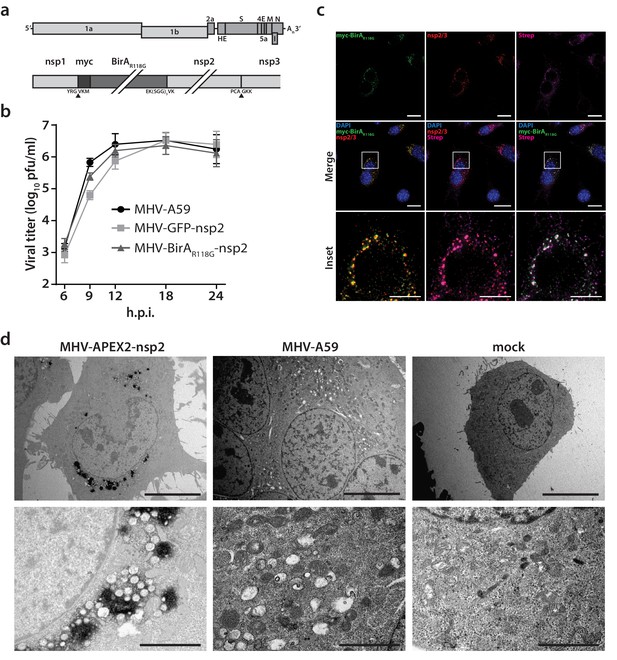
Characterization of the recombinant MHV-BirAR118G-nsp2.
(a) Genome organization of recombinant MHV-BirAR118G-nsp2. The positive-sense RNA genome of MHV contains a 5’ cap and a 3’ poly(A) tail. ORF1a and ORF1b encode the viral replication and transcription complex (nsp1-16). myc-BirAR118G was inserted as an N-terminal fusion with nsp2 within ORF1a. The cleavage site between nsp1 and myc-BirAR118G was retained (black arrow) while a deleted cleavage site between BirAR118G and nsp2 ensured the release of a BirAR118G-nsp2 fusion protein from the pp1a polyprotein. The cleavage site between nsp2 and nsp3 was also retained (grey arrow). (b) Viral replication kinetics of recombinant MHV-BirAR118G-nsp2 were compared to wild-type MHV-A59 and recombinant MHV-GFP-nsp2. Murine L929 fibroblasts were infected at a multiplicity of infection (MOI) of 1 plaque forming unit (pfu) per cell. Viral supernatants were collected at the indicated time points, titrated by plaque assay and expressed in pfu per ml. Data points represent the mean and SEM of three independent experiments, each performed in quadruplicate. (c) Immunofluorescence analysis of MHV-BirAR118G-nsp2-mediated biotinylation of RTC-proximal factors. L929 cells were infected with MHV-BirAR118G-nsp2 (MOI = 1) in medium supplemented with 67 µM biotin. Cells were fixed 15 hr post infection (h.p.i.) and processed for immunofluorescence analysis with antibodies directed against the BirAR118G (anti-myc), the viral replicase (anti-nsp2/3) and biotinylated factors (streptavidin). Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Scale bars: 20 µm; insets 5 µm. (d) Ultrastructural analysis of MHV-APEX2-nsp2 infection. L929 cells were infected with MHV-APEX2-nsp2 and MHV-A59 (MOI = 2), or mock infected. At 10 h.p.i., cells were fixed, stained with DAB and processed for electron microscopy investigations. Representative low (scale bar: 10 µm) and high magnifications (scale bar: 2 µm) are displayed.
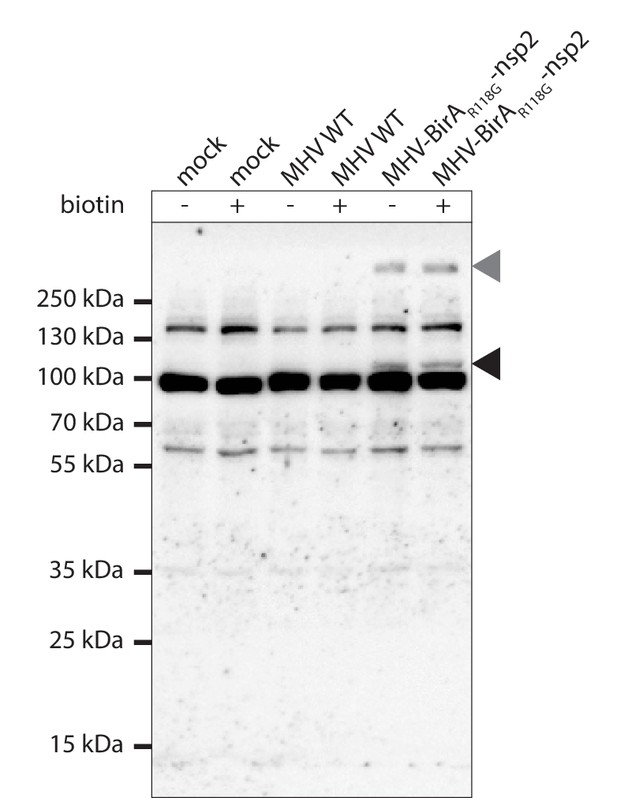
Western blot detection of BirAR118G-nsp2.
MHV-BirAR118G-nsp2, MHV-A59- or non-infected L929 fibroblasts were cultured in medium or in medium supplemented with 67 µM biotin for 15 hr. Lysates were separated by SDS-PAGE and western blots were probed using antibodies recognizing the N-terminal myc-tag of BirAR118G-nsp2. The black arrow indicates the fusion protein corresponding to the predicted molecular weight (102.2 kDa). High molecular weight proteins probably representing viral polyprotein precursors (grey arrow) were also specifically detected in MHV-BirAR118G-nsp2 –derived samples.
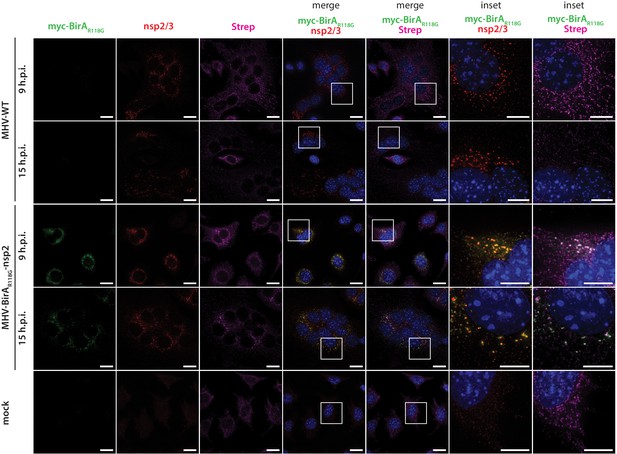
Immunofluorescence analysis of MHV-BirAR118G-nsp2-mediated biotinylation.
MHV-BirAR118G-nsp2, MHV-A59- or non-infected L929 fibroblasts were cultured in medium supplemented with 67 µM biotin. Cells were fixed 9 and 15 hr post infection (h.p.i.) and processed for immunofluorescence analysis with antibodies directed against the BirAR118G (anti-myc), the viral replicase (anti-nsp2/3) and biotinylated factors (streptavidin). Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Scale bars: 10 µm; insets 5 µm.

Immunofluorescence analysis of the MHV-BirAR118G-nsp2 RTC and BirAR118G-nsp2 –mediated biotinylataion.
MHV-BirAR118G-nsp2, MHV-A59- or non-infected L929 fibroblasts were cultured in medium supplemented with 67 µM biotin. Cells were fixed 12 hr post-infection (h.p.i.) and processed for immunofluorescence analysis with antibodies directed against the BirAR118G (anti-myc), the viral replicase (anti-nsp2/3, nsp8) and biotinylated factors (streptavidin). Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Scale bars: 10 µm; insets 5 µm.
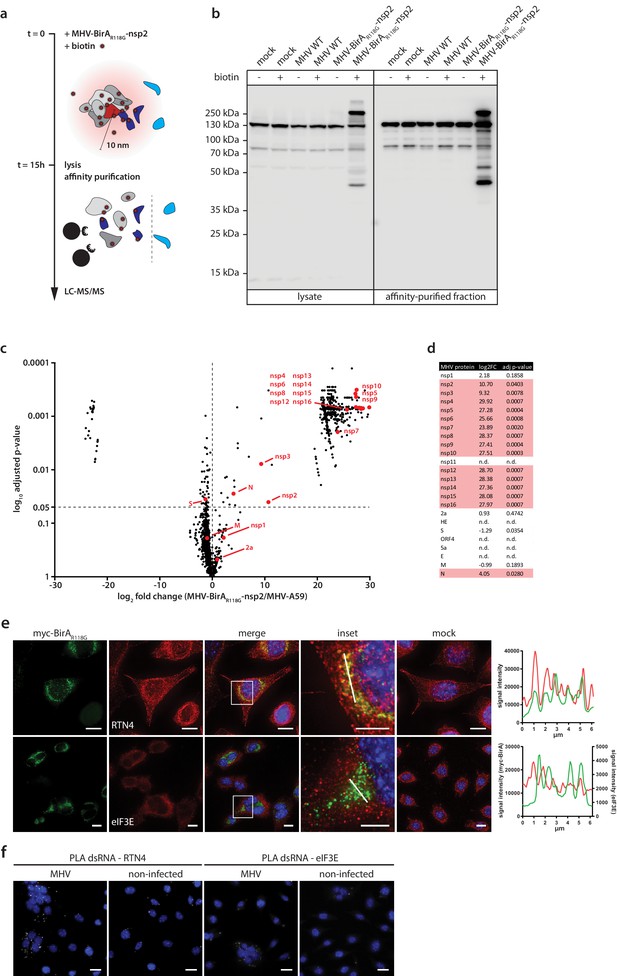
Determination of the coronavirus RTC-proximal proteome.
(a) Schematic overview of the BirAR118G-mediated proximity biotinylation assay using MHV-BirAR118G-nsp2. (b) Western blot analysis of MHV-BirAR118G-nsp2-infected L929 cells. L929 cells were infected with MHV-BirAR118G-nsp2, MHV-A59 or non-infected in medium with and without supplementation of 67 µM biotin. Cells were lysed 15 h.p.i. and biotinylated factors were subjected to affinity purification using streptavidin-coupled magnetic beads. Total cell lysates and affinity-purified fractions were separated by SDS-PAGE and analysed by western blot probed with horse radish peroxidase (HRP)-coupled Streptavidin. (c) Host and viral factors identified by LC-MS/MS. 4*107 L929 cells were infected with MHV-BirAR118G-nsp2 or MHV-A59 in medium supplemented with 67 µM biotin. 15 h.p.i., lysates were affinity purified and LC-MS/MS was performed from in-gel digested samples. MS identification of biotinylated proteins was performed in three independent biological replicates. Spectral interpretation was performed against a Mus musculus and MHV database and log2-transformed LFQ levels (x-axis) were used to determine significant differences in protein enrichment between sample groups (Student's T-test, y-axis). Identified cellular proteins are displayed as black dots, MHV proteins are highlighted in red (nsp: non-structural protein, N: nucleocapsid, S: spike, M: membrane, 2a: accessory protein 2a). (d) Summary of viral proteins identified by LC-MS/MS. nsp2-10, nsp12-16, and nucleocapsid were significantly enriched in fractions derived from MHV-BirAR118G-nsp2-infected cells whereas nsp1, nsp11, structural proteins spike (S), envelope (E) and membrane proteins (M) as well as all accessory proteins (NS2a, HE, ORF4, ORF5a) were either not significantly enriched or not detected. (e,f) Immunofluorescence analysis of RTC-proximal cellular factors. L929 cells were seeded on coverslips, infected with MHV-BirAR118G-nsp2 (e) or MHV-A59 (f), fixed at 9 h.p.i. and processed for immunofluorescence using anti-myc, anti-RTN4 and anti-eIF3E antibodies (e) or anti-dsRNA, anti-RTN4 and anti-eIF3E antibodies (f). Secondary fluorophore-coupled antibodies were used to detect the viral replicase and endogenous levels of RTN4 and eIF3E (e). Scale bars: 10 µm; insets 5 µm. Proximity ligations were performed using Duolink In Situ detection reagents (f). Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Intensity profiles highlighted in the magnified regions are shown. Scale bars: 20 µm (insets 5 µm).
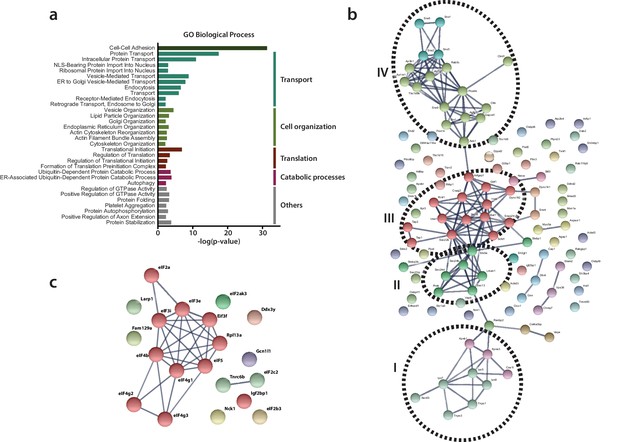
Functional classification of RTC-proximal host factors.
(a) Gene Ontology enrichment analysis of RTC-proximal cellular factors. 32 terms were highly significant (p-value < 0.005) and were assigned to five broad functional categories: cell-cell adhesion, transport, cell organization, translation, catabolic processes. (b–c) STRING protein interaction network analysis of the categories ‘transport’ (b) and ‘translation’ (c). The nodes represent RTC-proximal host proteins and the edges represent the interactions, either direct (physical) or indirect (functional), between two proteins in the network. Cellular proteins assigned to the ‘transport’ category separated into four distinct interaction clusters. I: protein transport, II: COPII anterograde transport, III: COPI retrograde transport, IV: clathrin-mediated transport.
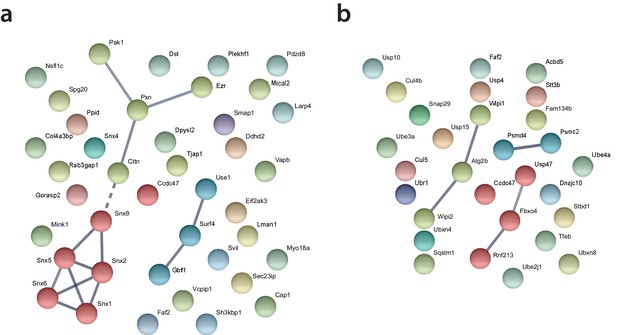
STRING protein interaction network analysis of the categories ‘cell organization’ (a) and ‘catabolic processes’ (b).
The nodes represent RTC-proximal host proteins and the edges represent the interactions, either direct (physical) or indirect (functional), between two proteins in the network.
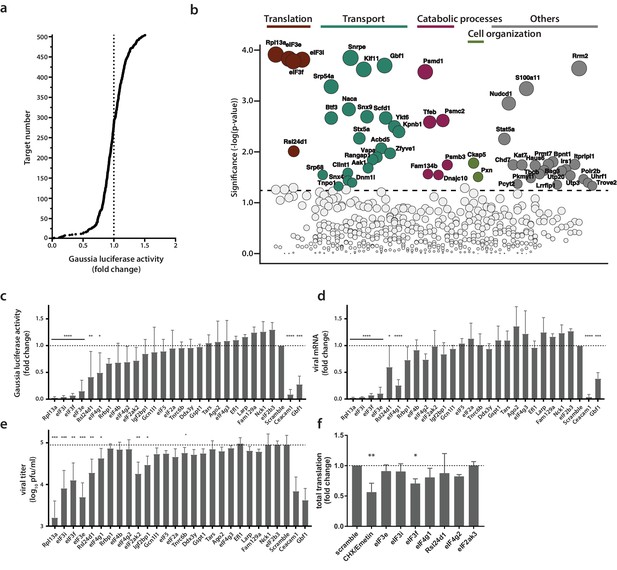
Identification of proviral factors within the coronavirus RTC microenvironment.
(a) Impact of siRNA-silencing of RTC-proximal cellular proteins on viral replication. L929 fibroblasts were reverse-transfected with siRNAs (10 nM) for 48 hr before being infected with MHV-Gluc (MOI = 0.05, n = 4). Replication was assessed by virus-mediated Gaussia luciferase expression at 15 h.p.i. and was normalized to levels of viral replication in cells targeted by scrambled siRNA controls. Target proteins to the left of the dashed line represent RTC-proximal factors whose silencing decreased viral replication. (b) Bubble plot illustrating host proteins that significantly impact MHV replication. Bubble size is proportional to the level of viral replication impairment. Colors correspond to the functional categories highlighted in Figure 3. Light grey bubbles (below the dashed line) represent host proteins that did not significantly impact MHV replication (p-value > 0.05). (c, d, e, f) Silencing of RTC-proximal components of the cellular translation machinery. Upon 48 hr siRNA silencing of factors assigned to the category ‘translation’ (Figure 3), L929 fibroblasts were infected with MHV-Gluc (MOI = 0.05, n = 3). Luciferase activity (c), cell-associated viral RNA levels (d) and viral titers (e) were assessed at 12 h.p.i.. (f) Western blot quantification of total cellular translation following silencing of a subset of the host translation apparatus. Upon 48 hr siRNA-silencing, L929 fibroblasts were pulsed with 3 µM puromycin for 60 min. Control cells were treated, prior to puromycin incubation, with 355 µM cycloheximide and 208 µM Emetin for 30 min to block protein synthesis. Cell lysates were separated by SDS-PAGE and Western blots were probed using anti-puromycin antibodies to assess puromycin incorporation into polypeptides and normalized to actin levels. Error bars represent the mean ± standard deviation, where * is p ≤ 0.05, ** is p ≤ 0.005, *** is p ≤ 0.0005 and **** is p < 0.0001.
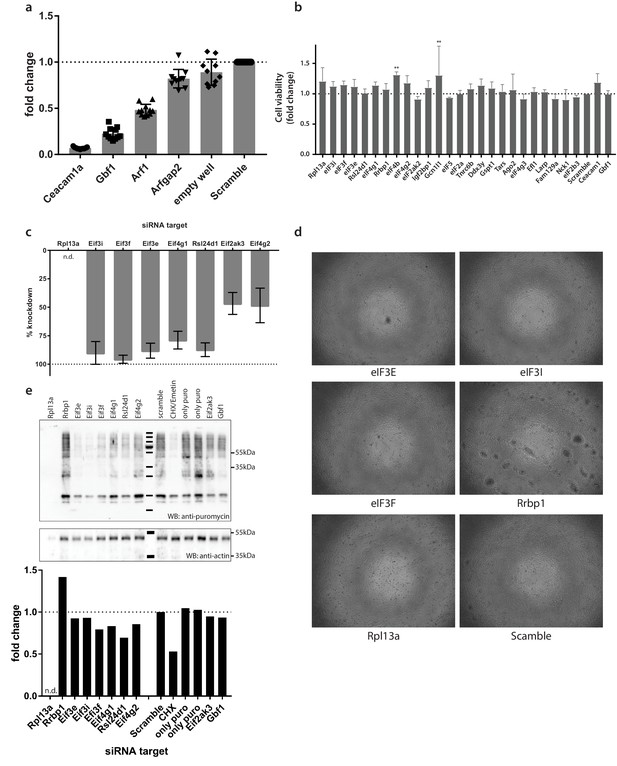
(a) siRNA controls contained in each 96-well plate during siRNA-silencing of the RTC-proximal library.
Controls included the established factors such as MHV entry receptor (Ceacam1a), Gbf1, Arf1. Arfgap2 was found to moderately affect MHV replication during pilot experiments and was included to cover the entire inhibitory range. (b) Cell viability following 48 hr siRNA-silencing of components of the cellular translation machinery. (c) Expression levels of Rpl13a, eIF3E, eIF3I, eIF3F, eIF4G1, eIF4G2, eIF2ak3, Rsl24d1 following siRNA knockdown compared to expression levels in cells treated with non-targetting siRNA. (d) Visual inspection of L929 treated with siRNA targetting eIF3E, eIF3I, eIF3F, Rrbp1, Rpl13a, non-targetting siRNA (scramble). Note that RNA silencing (b) and translation activity (c) in Rpl13a-silenced cells could not be assessed, likely due to cytotoxicity observed by visual inspection of cells. (e) Western blot and western blot analysis of total cellular translation. Upon 48 hr siRNA-silencing, L929 fibroblasts were pulsed with 3 µM puromycin for 60 min. Control cells were treated, prior to puromycin incubation, with 355 µM cycloheximide and 208 µM Emetin for 30 min to block protein synthesis. Western blots were probed using anti-puromycin antibodies to assess puromycin incorporation into polypeptides and normalized to actin levels. Error bars represent the mean ±standard deviation of three independent experiments, where * is ** is p ≤ 0.005.
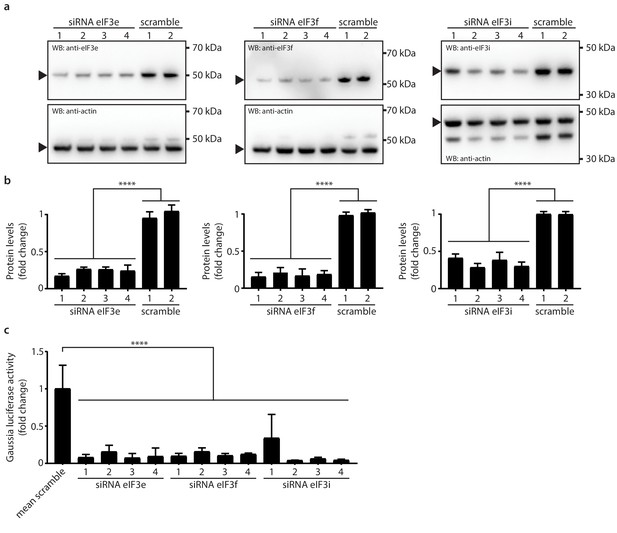
Knockdown of eIF3e, eIF3f and eIF3i impact MHV replication.
(a) siRNA silencing and western blot assessment of eIF3e, eIF3f and eIF3i protein levels. 10 nM individual target-specific siRNA were transfected in L929 for 48 hr. Lysates were separated by SDS-PAGE and western blots were probed using antibodies directed against eIF3e, eIF3f and eIF3i. The same membranes were subsequently washed, and actin, which was used to normalize protein levels, was detected using a conjugated antibody. Arrows indicate proteins bands of interest. (b) protein quantification of eIF3e, eIF3f and eIF3i upon siRNA-mediated knockdown using single target-specific siRNAs (n = 3). Error bars represent the mean ± standard deviation, where **** is p < 0.0001. (c) L929 fibroblasts were infected with MHV-Gluc (MOI = 0.05, n = 3) for 12 hr upon a 48 hr knockdown of eIF3e, eIF3f and eIF3i using single target-specific siRNAs. Luciferase counts reflecting viral replication were normalized to scrambled non-targeting siRNA controls. Error bars represent the mean ± standard deviation, where **** is p < 0.0001.
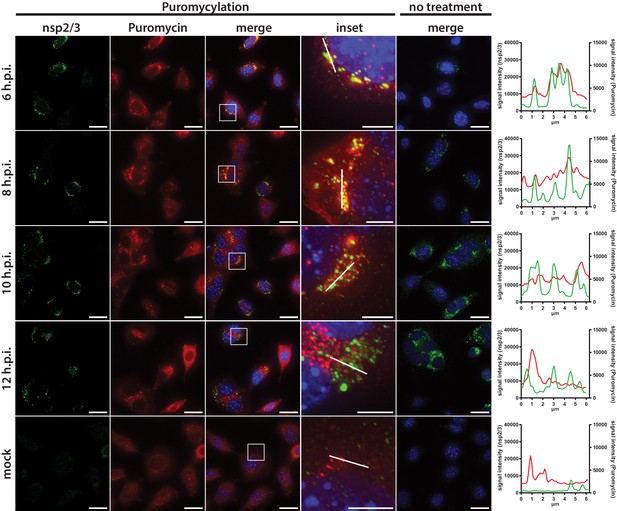
Active translation near sites of MHV mRNA synthesis.
Visualization of active translation in MHV-infected L929 fibroblasts. Cells infected with MHV-A59 (MOI = 1) or non-infected cells were cultured for 6, 8, 10 and 12 hr and pulsed with cycloheximide, emetine and puromycin for 5 min to label translating ribosomes. All cells, including non-treated control infections, were subjected to a coextraction/fixation procedure to remove free puromycin. Cells were labeled using anti-nsp2/3 antiserum and anti-puromycin antibodies. Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Note the gradual decrease of overlap between the viral replication and actively translating ribosomes highlighted in the intensity profiles. Scale bar: 20 µm; insets 5 µm.
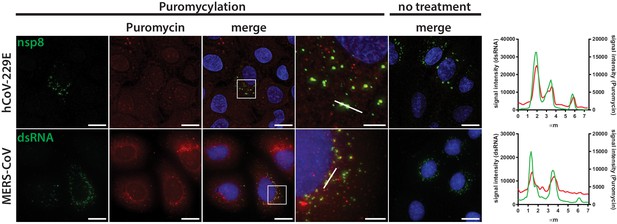
Active translation near sites of HCoV-229E and MERS-CoV mRNA synthesis.
Visualization of active translation during HCoV-229E and MERS-CoV infections. Huh7 cells were infected with HCoV-229E and MERS-CoV (MOI = 1) for 12 hr and 6 hr, respectively. Cells were pulsed with cycloheximide, emetine and puromycin for 5 min to label translating ribosomes and subjected to a coextraction/fixation procedure to remove free puromycin. Non-infected and/or non-pulsed cells were used as control. Cells were labelled using anti-nsp8 (HCoV-229E) or dsRNA (MERS-CoV) and anti-puromycin antibodies. Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Intensity profiles in magnified regions are shown. Scale bar: 20 µm; insets 5 µm.
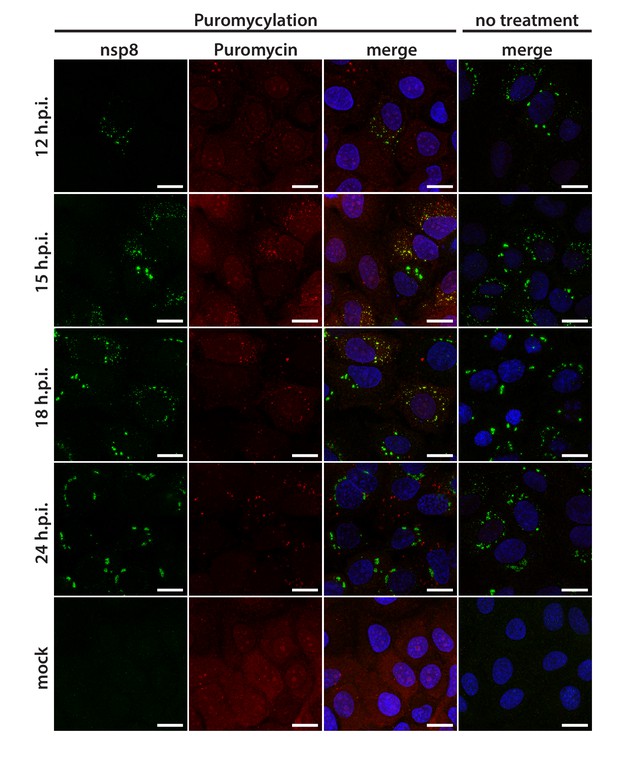
Visualization of active translation during HCoV-229E infections.
Huh7 cells were infected with HCoV-229E (MOI = 1) for 12, 15, 18, 24 hr. Cells were pulsed with cycloheximide, emetine and puromycin for 5 min to label translating ribosomes and subjected to a coextraction/fixation procedure to remove free puromycin. Non-infected and/or non-pulsed cells were used as control. Cells were labeled using anti-nsp8 (HCoV-229E) and anti-puromycin antibodies. Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Scale bar: 20 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | BirAR118G | PMCID: 3308701 | ||
| Strain, strain background (mouse hepatitis virus) | MHV-A59 | PMID: 15709029 | ||
| Strain, strain background (mouse hepatitis virus) | MHV-Gluc | PMID: 24874215 | ||
| Strain, strain background (mouse hepatitis virus) | MHV-BirAR118G-nsp2 | This study | ||
| Strain, strain background (human coronavirus) | HCoV-229E | PMID: 19057873 | ||
| Strain, strain background (middle east respiratory syndrome coronavirus) | MERS-CoV | PMID: 23170002; 23078800 | ||
| Cell line (Mus musculus) | L929 | Sigma | 85011425 | |
| Cell line (Mus musculus) | 17Cl1 | Gift from S.G. Sawicki | PMC422565 | |
| Cell line (Homo sapiens) | Huh7 | Gift from V. Lohmann | CVCL_0336 | |
| Cell line (african green monkey) | Vero B4 | Gift from M. Müller | CVCL_1912 | |
| Antibody | anti-dsRNA J2 (mouse monoclonal IgG2a, kappa chain) | English and Scientific Consulting | Product No: 10010500 | 1:200 |
| Antibody | anti-myc (mouse monoclonal) | Cell signalling | 2276 | 1:8000 (IF); 1:1000 (WB) |
| Antibody | Anti-Nogo A + B (rabbit polyclonal) | Abcam | Product No : ab47085 | 1:200 |
| Antibody | Anti-EIF3E (rabbit polyclonal) | Sigma | HPA023973 | 1:100 (IF); 1:300 (WB) |
| Antibody | Anti-EIF3F (rabbit polyclonal) | Abcam | ab176853 | 1:3000 (WB) |
| Antibody | Anti-EIF3I (rabbit polyclonal) | Sigma | HPA029939 | 1:500 (WB) |
| Antibody | Anti-Puromycin(mouse monoclonal IgG2a,κ) | Merk Millipore | MABE343 | 1:10000 |
| Antibody | Anti-MHV nsp2/3 (rabbit polyclonal) | Gift from S. Baker | PMID: 9514967 | 1:200 |
| Antibody | Anti-MHV nsp8 (rabbit polyclonal) | Gift from S. Baker | PMID: 11907209 | 1:400 |
| Antibody | Anti-229E-nsp8 (rabbit polyclonal) | Gift from J Ziebuhr | PMID: 9847320 | 1:200 |
| Antibody | donkey anti-mouse 488 | Jackson ImmunoResearch | 715-545-150 | 1:400 |
| Antibody | donkey anti-rabbit 594 | Jackson ImmunoResearch | 711-585-152 | 1:400 |
| Antibody | donkey anti-rabbit 647 | Jackson ImmunoResearch | 711-605-152 | 1:400 |
| Antibody | donkey anti-rabbit HRP | Jackson ImmunoResearch | 711-035-152 | 1:10000 |
| Antibody | donkey anti-mouse HRP | Jackson ImmunoResearch | 715-035-151 | 1:5000 |
| Antibody | anti-actin HRP (mouse monoclonal) | Sigma | A3854 | 1:25000-1:50000 |
| Sequence-based reagent | On-Target Plus CherryPick siRNA Library | Horizon Discovery Ltd. | ||
| Commercial assay or kit | Pierce Gaussia Luciferase Glow Assay Kit | ThermoFisher Scientific | 16160 | |
| Commercial assay or kit | CytoTox 96 Non-Radioactive Cytotoxicity Assay | Promega | G1780 | |
| Commercial assay or kit | Viromer Green | Lipocalyx | VG-01LB-00 | |
| Commercial assay or kit | Dynabeads MyOne Streptavidin C1 | ThermoFisher Scientific | 65001 | |
| Chemical compound, drug | Puromycin | Sigma | P9620 | |
| Chemical compound, drug | Cycloheximide | Sigma | C7698 | |
| Chemical compound, drug | Emetin | Sigma | E2375 | |
| Chemical compound, drug | Biotin | Sigma | B4501 |
Additional files
-
Supplementary file 1
Mass spectrometry data.
- https://doi.org/10.7554/eLife.42037.016
-
Supplementary file 2
Gene Ontology enrichment analysis and STRING functional protein association association network analysis.
- https://doi.org/10.7554/eLife.42037.017
-
Supplementary file 3
siRNA-mediated knockdown screen.
- https://doi.org/10.7554/eLife.42037.018
-
Supplementary file 4
Primer and probes used in RT-qPCR.
- https://doi.org/10.7554/eLife.42037.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42037.020





