The ULK1-FBXW5-SEC23B nexus controls autophagy
Figures
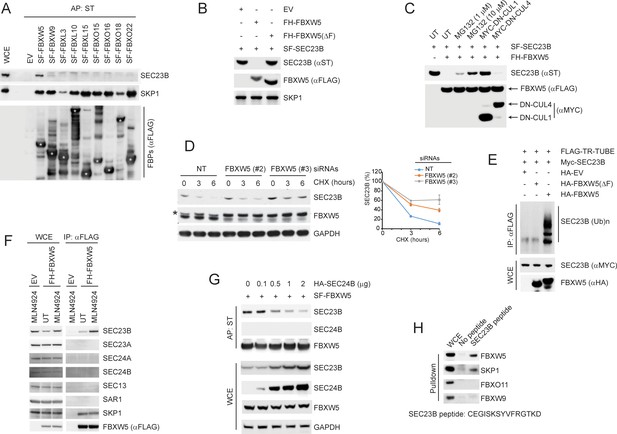
SCFFBXW5 interacts with SEC23B and targets it for ubiquitylation and proteasome-mediated degradation.
(A) HEK293T cells were transfected with either an empty vector (EV) or the indicated Streptag-FLAG-tagged (SF) F-box proteins (FBPs). Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated. (WCE, whole cell extracts). The white asterisks indicate individual F-box proteins. (B) HEK293T cells were transfected with an EV, FLAG-HA-tagged FBXW5 (FH-FBXW5), or FH-FBXW5(ΔF) together with SF-tagged SEC23B. Twenty-four hours after transfection, cells were harvested for immunoblotting. (C) HEK293T cells were transfected with FH-FBXW5 and SF-SEC23B in combination with either an EV, MYC-tagged DN-CUL1, or MYC-tagged DN-CUL4 as indicated. Twenty-four hours after transfection, cells were either left untreated (UT) or treated with MG132 for 6 hr, and finally harvested for immunoblotting. (D) U-2OS cells were transfected with either a non-targeting siRNA oligo (NT) or two different FBXW5 siRNA oligos (individually). Seventy-two hours after siRNA transfection, cells were treated with cycloheximide (CHX) for the indicated times and harvested for immunoblotting. The asterisk indicates a nonspecific band. The graph shows the quantification of SEC23B levels from three independent experiments. Error bars indicate standard deviation. (E) HEK293T cells were transfected with an EV, HA-tagged FBXW5, or HA-tagged FBXW5(ΔF) together with MYC-tagged SEC23B and FLAG-TR-TUBE cDNA as indicated. WCLs were immunoprecipitated (IP) with anti-FLAG resin and immunoblotted as indicated. The line on the right marks a ladder of bands corresponding to poly-ubiquitylated SEC23B. (F) HEK293T cells were transfected with either an EV or FH-FBXW5. Twenty-four hours after transfection, cells were either left untreated (UT) or treated with MLN4924 for 4 hr before harvesting them for immunoprecipitation (IP) with FLAG beads and immunoblotting as indicated. (G) HEK293T cells were transfected with SF-FBXW5 and increasing amounts of HA-tagged SEC24B as indicated. Twenty-four hours after transfection, cells were harvested for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated. (H) WCEs from HEK293T cells were incubated with either unconjugated beads or beads coupled to a SEC23B peptide (a.a. 180–194, CEGISKSYVFRGTKD). Beads were washed with lysis buffer and bound proteins were eluted and subjected to SDS-PAGE and immunoblotting.
-
Figure 1—source data 1
Source data for Figure 1B.
- https://doi.org/10.7554/eLife.42253.004
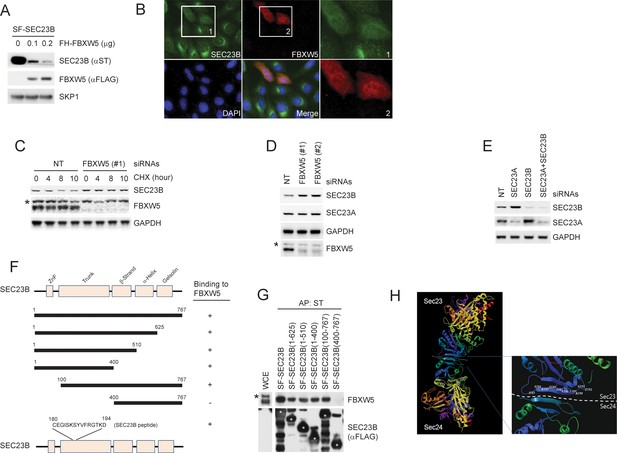
FBXW5 interacts with and promotes the degradation of SEC23B.
(A) HEK293T cells were transfected with Streptag-FLAG-tagged SEC23B (SF-SEC23B) and increasing amounts of FLAG-HA-tagged FBXW5 (FH-FBXW5). Twenty-four hours after transfection, cells were harvested for immunoblotting as indicated. (B) U-2OS cells stable expressing FLAG-tagged SEC23B were transfected with HA-tagged FBXW5. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde in PBS. An anti-FLAG antibody and an anti-HA antibody were used for the detection of SEC23B and FBXW5, respectively. Boxed images were enlarged for clarity. (C) RPE1-hTERT cells were transfected with either a non-targeting siRNA oligo (NT) or a FBXW5 siRNA oligo. Seventy-two hours after siRNA transfection, cells were treated with cycloheximide (CHX) for the indicated times and harvested for immunoblotting as indicated. The asterisk indicates a nonspecific band. (D) RPE1-hTERT cells were transfected with either a non-targeting siRNA oligo (NT) or two different FBXW5 siRNA oligos (each individually). Seventy-two hours after siRNA transfection, cells were harvested for immunoblotting as indicated. The asterisk indicates a nonspecific band recognized by the anti-FBXW5 antibody. (E) RPE1-hTERT cells were transfected with siRNA oligos to the indicated mRNAs. Seventy-two hours after siRNA transfection, cells were harvested for immunoblotting. (F) Schematic view of the SEC23B mutants used in Figure 1—figure supplement 1G and the SEC23B degron peptide. (G) HEK293T cells were transfected with the truncated mutants shown in Figure 1—figure supplement 1F. Twenty-four hours after transfection, cells were harvested for affinity-purification (AP) with Streptactin (ST) beads followed by immunoblotting as indicated. The black asterisk indicates a nonspecific band. The white asterisks indicate bands corresponding to SEC23B or SEC23B mutants. (H) Crystal structure showing the position of Serine 186 of SEC23. Images (PDB: 3EFO; Mancias and Goldberg, 2008) were created by MolBrowser (Molsoft). The dashed line indicates the interface between SEC23 and SEC24 where serine 186 resides.
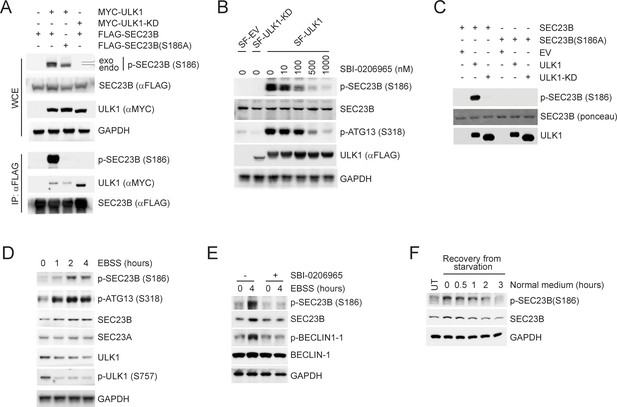
ULK1 phosphorylates SEC23B on Serine 186.
(A) HEK293T cells were transfected with either FLAG-tagged SEC23B or FLAG-tagged SEC23B(S186A) in combination with MYC-tagged ULK1 or MYC-tagged ULK1-KD as indicated. Twenty-four hours after transfection, cells were harvested for immunoprecipitation (IP) and immunoblotting. exo and endo indicate the exogenous and endogenous SEC23B, respectively. (B) HEK293T cells were transfected with the SF-ULK1 or SF- ULK1-KD as indicated. Twenty-four hours after transfection, cells were treated with various doses of SBI-0206965 (an ULK1 inhibitor) for 4 hr before harvesting them for immunoblotting. (C) In vitro kinase assays were performed using purified SEC23B (wild-type or the S186A mutant) and ULK1 (wild-type or a kinase-dead mutant) as substrate and kinase, respectively. Purified SEC23B and ULK1 proteins were prepared by immunoprecipitation (followed by elution) from extracts of HEK293T cells transfected with each corresponding plasmid. (D) HEK293T cells were nutrient-starved with EBSS for the indicated times and harvested for immunoblotting. (E) HEK293T cells were nutrient-starved with EBSS for the indicated times (in the presence or absence of SBI-0206965) and harvested for immunoblotting at the indicated times. (F) HEK293T cells were recovered from nutrient-starvation (EBSS for 4 hr) for the indicated times and harvested for immunoblotting.
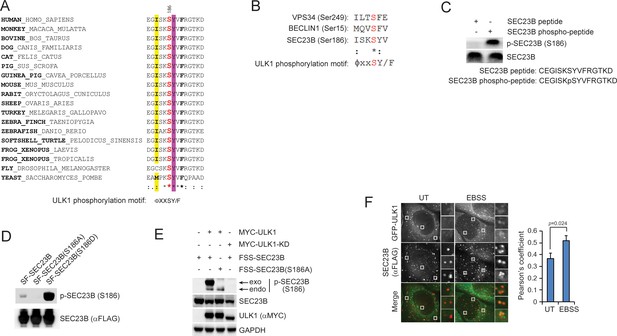
Characterization of the phospho-SEC23B (Ser186) antibody and the phospho-mimetic SEC23B mutant.
(A) An ULK1 phosphorylation motif (ΦxxSY/F)(Egan et al., 2015) is highly conserved throughout evolution in the FBXW5-binding region of SEC23B. Φ, hydrophobic amino acids. (B) Sequence alignment of the previously characterized ULK1-substrates with SEC23B. (C) A non-phosphorylated SEC23B peptide and an equivalent phospho-peptide (sequences are indicated below the panels) were separated by SDS-PAGE and subjected to immunoblotting either with a phospho-specific SEC23B (Ser186) antibody or with an anti-SEC23B antibody reactive against both phosphorylated and non-phosphorylated species of SEC23B. (D) HEK239T cells were transfected with either SF- tagged SEC23B or the indicated SF-tagged SEC23B mutants. Twenty-four hours after transfection, cells were immunoblotted as indicated. (E) HEK293T cells were transfected with either FLAG-Streptag-Streptag-tagged SEC23B or FLAG-Streptag-Streptag-tagged SEC23B(S186A) in combination with MYC-tagged ULK1 or MYC-tagged ULK1-KD as indicated. Twenty-four hours after transfection, cells were harvested for immunoblotting as indicated. exo and endo indicate the exogenous and endogenous SEC23B, respectively. (F) U-2OS cells stably expressing GFP-ULK1 were transfected with FLAG-HA-tagged SEC23B. Twenty-four hours after transfection, cells were fixed for immunofluorescence as indicated. Images were analysed by ImageJ with at least 100 cells counted per sample. Quantification of SEC23B overlapping with GFP-ULK1 was performed using the Pearson's correlation coefficient. The data are presented as mean ±SD (right panel). Scale bar, 10 μm.
-
Figure 2—figure supplement 1—source data 1
Source data for panel F.
- https://doi.org/10.7554/eLife.42253.007
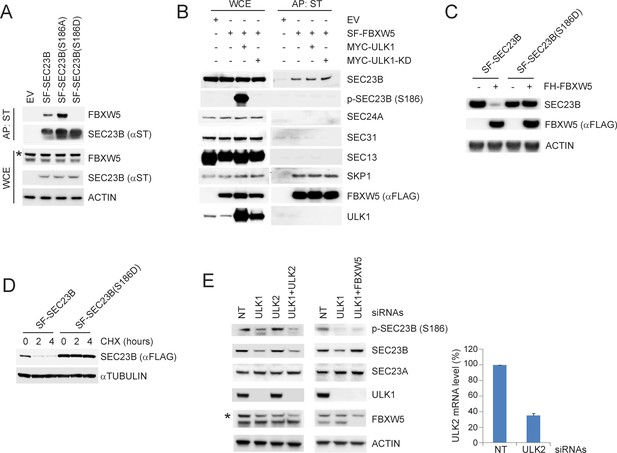
ULK1-mediated phosphorylation of SEC23B on S186 prevents the FBXW5-dependent degradation of SEC23B.
(A) HEK293T cells were transfected with either EV, SF-SEC23B, or the indicated SF-SEC23B mutants. Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting. The asterisk indicates a nonspecific band. (B) HEK293T cells were transfected with SF-FBXW5 in combination with either MYC-tagged ULK1 or MYC-tagged ULK1-KD. Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated. (C) HEK293T cells were transfected with FH-FBXW5 in combination with SF-SEC23B or SF-SEC23B(S186D). Twenty-four hours after transfection, cells were harvested for immunoblotting. (D) U-2OS cells stably infected with viruses expressing either SEC23B or SEC23B(S186D) were treated with cycloheximide for the indicated times. The cells were then harvested for immunoblotting. (E) RPE1-hTERT cells were transfected with siRNAs against the indicated mRNAs. Sixty-eight hours after transfection, cells were nutrient-starved with EBSS for 4 hr and harvested for immunoblotting (left panel) and real-time PCR using ULK2 and GAPDH primers (right panel). The asterisk indicates the nonspecific band.
-
Figure 3—source data 1
Source data for Figure 3E.
- https://doi.org/10.7554/eLife.42253.010
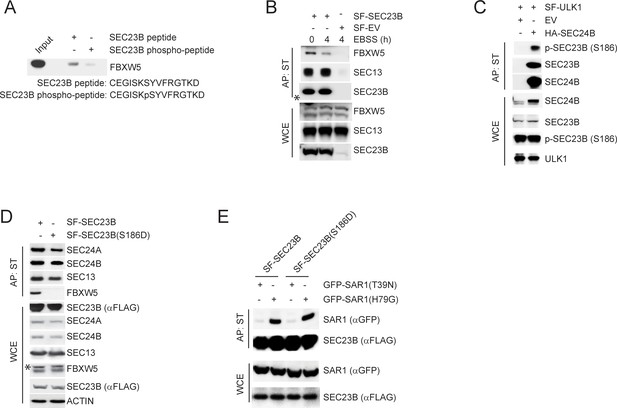
ULK1-mediated phosphorylation of SEC23B on S186 prevents the FBXW5-dependent degradation of SEC23B.
(A) A non-phosphorylated SEC23B peptide and an equivalent phospho-peptide (sequences are indicated below the panels) were conjugated to beads and used in an in vitro binding assay with FBXW5 that was in vitro transcribed/translated using a rabbit reticulocyte system. Bound FBXW5 was detected by immunoblotting. (B) HEK293T cells were transfected with either a Streptag-FLAG empty vector (SF-EV) or Streptag-FLAG-tagged SEC23B (SF-SEC23B). Twenty-four hours after transfection, cells were nutrient-starved with EBSS for the indicated times and treated with MLN4924 for 4 hr before harvesting them for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated. (WCE, whole cell extracts). The white asterisk indicates a non-specific band. (C) HEK239T cells were transfected with an empty vector (EV) or HA-tagged SEC24B together with Streptag-FLAG tagged ULK1 (SF-ULK1) as indicated. Twenty-four hours after transfection, cells were harvested for immunoprecipitation (IP) with HA beads and immunoblotting as indicated. (WCE, whole cell extracts). (D) HEK239T cells were transfected with either Streptag-FLAG tagged wild-type SEC23B or the phospho-mimetic SEC23B(S186D) mutant (also Streptag-FLAG tagged). Twenty-four hours after transfection, cells were harvested for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated. (WCE, whole cell extracts). (E) HEK239T cells were transfected with either Streptag-FLAG tagged SEC23B (SF-SEC23B) or SF-SEC23B(S186D), in combination with the indicated GFP-tagged SAR1 mutants. Twenty-four hours after transfection, cells were harvested for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting as indicated.
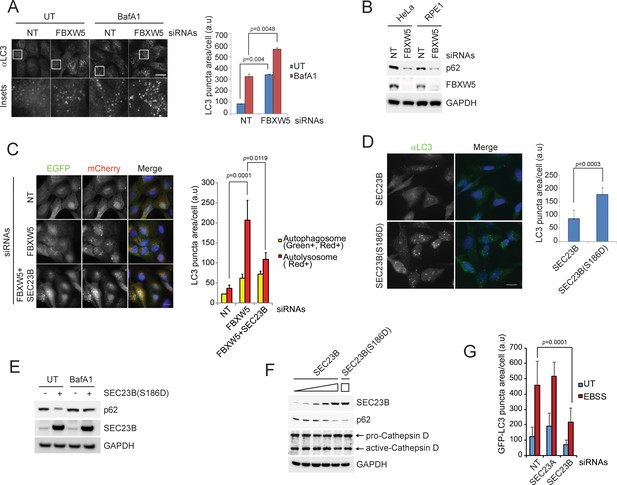
The FBXW5-mediated degradation of SEC23B limits the autophagic flux in the presence of nutrients.
(A) RPE1-hTERT cells were transfected with a non-targeting (NT) oligo or a FBXW5-targeting siRNA oligo. Forty-eight hours after transfection, cells were re-plated onto coverglass for immunofluorescence with an anti-LC3 antibody. Where indicated, cells were treated with Bafilomycin A1 (BafA1) for 4 hr before fixation. Images of endogenous LC3 puncta underwent automated processing with at least 300 cells counted per sample. Because in several images LC3·puncta were too close to be distinguished, we adopted LC3 puncta area as a criterion for our analysis. The data are presented as mean ±SD (right panel). Scale bar, 10 μm. (B) HeLa and RPE1-hTERT cells were transfected with the indicated siRNAs. Seventy-two hours after transfection, the cells were harvested for immunoblotting. (C) U-2OS cells stably expressing tandem fluorescent-tagged LC3 (pBabe-mCherry-EGFP-LC3) were transfected with a NT oligo or a FBXW5 siRNA oligo, alone or in combination with a SEC23B-targeting siRNA oligo as indicated. Forty-eight hours after transfection, cells were replated onto coverglass, followed by fixation twenty-four hours after replating. Images of mCherry-EGFP-LC3 puncta underwent automated processing with at least 100 cells counted per sample. The data are presented as mean ±SD (right panel). The yellow and red bars represent green +red double positive LC3 puncta (autophagosome) and red only positive LC3 puncta (autolysosome), respectively. Scale bar, 10 μm. (D) U-2OS cells were infected with lentiviruses expressing either wild-type SEC23B or SEC23B(S186D). Twenty-four hours after infection, cells were fixed for immunofluorescence. Images of endogenous LC3 puncta underwent automated processing with at least 300 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm. (E) U-2OS cells were infected with lentivirus expressing SEC23B(S186D). Where indicated, forty-eight hours after infection, cells were treated with BafA1 prior to harvest and immunoblotting. (F) U-2OS cells were infected with the increasing amounts of lentivirus expressing SEC23B. Forty-eight hours after infection, cells were harvested for immunoblotting. (G) RPE1-hTERT cells stably expressing GFP-tagged LC3 were transfected with a NT oligo or the indicated siRNA oligos. Forty-eight hours after transfection, cells were replated onto coverglass, followed by treatment with EBSS for 1 hr and fixation. Images of GFP-LC3 puncta underwent automated processing with at least 300 cells counted per sample. The data are presented as mean ±SD.
-
Figure 4—source data 1
Source data for Figure 4A,C,D and G.
- https://doi.org/10.7554/eLife.42253.014
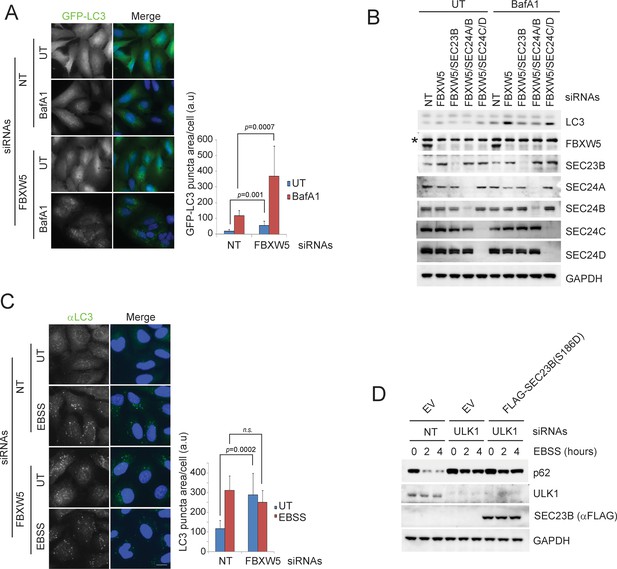
The FBXW5-mediated degradation of SEC23B limits the autophagic flux in the presence of nutrients.
(A) RPE1-hTERT cells stably expressing GFP-LC3 were transfected with either a non-targeting siRNA oligo (NT) or an siRNA oligo targeting FBXW5 RNA. Forty-eight hours after transfection, cells were replated onto coverglass. Where indicated, cells were treated with Bafilomycin A1 (BafA1) for 4 hr before fixation with cold methanol. Images of the GFP-LC3 puncta underwent automated processing with at least 300 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm. (B) RPE1-hTERT cells were transfected with the indicated siRNAs. Seventy-two hours after transfection, where indicated, cells were treated with Bafilomycin A1 (BafA1) for one hour prior to harvest for immunoblotting. (C) RPE1-hTERT cells stably expressing GFP-LC3 were transfected with either a non-targeting siRNA oligo (NT) or an siRNA oligo targeting FBXW5 RNA. Forty-eight hours after transfection, cells were replated onto coverglass for immunofluorescence with an anti-LC3 antibody. Cells were nutrient-starved with EBSS for 4 hr before fixation with cold methanol. Images of the LC3 puncta underwent automated processing with at least 300 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm. (D) U-2OS cells infected with either an empty virus (EV) or lentivirus expressing SEC23B(S186D) were transfected with the indicated siRNAs. Seventy-two hours after transfection, cells were treated with EBSS prior to harvest for immunoblotting.
-
Figure 4—figure supplement 1—source data 1
Source data for panels A and C.
- https://doi.org/10.7554/eLife.42253.013
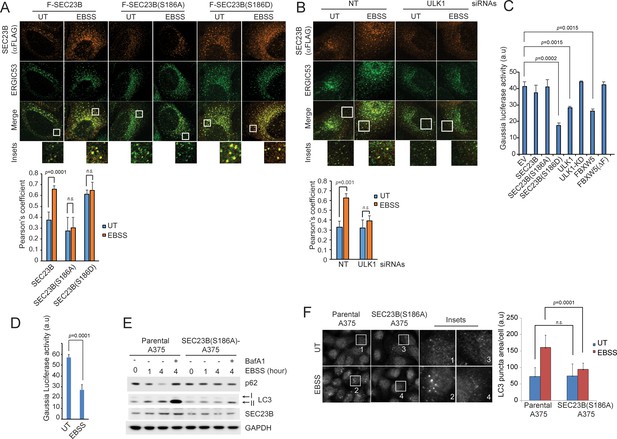
Ser186 in SEC23B is necessary for its localization to the ERGIC and an efficient autophagic response upon nutrient deprivation.
(A) U-2OS cells were transfected with either FLAG-HA-tagged wild-type SEC23B, SEC23B(S186A), or SEC23B(S186D). Twenty-four hours after transfection, cells were fixed for immunofluorescence as indicated. Images were analysed by ImageJ with at least 100 cells counted per sample. Quantification of SEC23B overlapping with ERGIC53 was performed using the Pearson's correlation coefficient. The data are presented as mean ±SD (bottom panel). Scale bar, 10 μm. (B) U-2OS cells stably expressing FLAG-HA-tagged SEC23B were transfected with ULK1 siRNAs. Seventy-two hours after transfection, cells were fixed for immunofluorescence as indicated. Images were analysed by ImageJ with at least 100 cells counted per sample. Quantification of SEC23B overlapping with ERGIC53 was performed using the Pearson's correlation coefficient. The data are presented as mean ±SD (bottom panel). Scale bar, 10 μm. (C) HEK239T cells were transfected with a plasmid expressing Gaussia luciferase in combination with the indicated constructs. Twenty-four hours after transfection, cells were replated onto 96-well plates. After another forty-eight hours, fresh media was added to the cells, and four hours after, the culture media were collected to measure Gaussia luciferase activity. The data are presented as mean ±SD of the Gaussia luciferase activity of triplicate samples. Expression of FBXW5 was used as a positive control since it results in the downregulation of SEC23B and, therefore, it is expected to inhibit trafficking. (D) A375 cells stably expressing Gaussia luciferase were plated onto 96-well plates. After forty-eight hours, either fresh media or EBSS was added to the cells and four hours after, the culture media were collected to measure Gaussia luciferase activity. The data are presented as mean ±SD of the Gaussia luciferase activity of triplicate samples. (E) A375 parental cells or SEC23B(S186A)-A357 knock-in cells were starved with EBSS for the indicated times (±BafA1) and harvested for immunoblotting as indicated. (F) A375 parental cells or SEC23B(S186A)-A357 knock-in cells were starved with EBSS and fixed for immunofluorescence. Images of endogenous LC3 puncta underwent automated processing with at least 100 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm.
-
Figure 5—source data 1
Source data for Figure 5B,C,D and F.
- https://doi.org/10.7554/eLife.42253.017
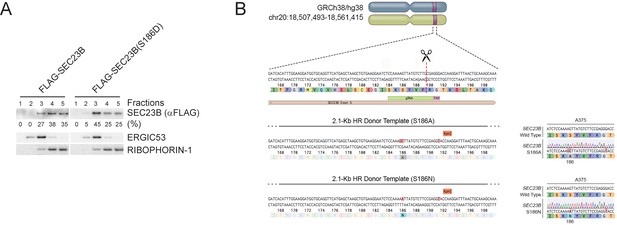
Ser186 in SEC23B is necessary for its localization to the ERGIC and an efficient autophagic response upon nutrient deprivation.
(A) HeLa cells were permeabilized, incubated with either purified recombinant FLAG-tagged SEC23B or purified recombinant FLAG-tagged SEC23B(S186D), and subjected to subcellular fractionation. Each fraction was subjected to immunoblotting with the indicated antibodies. ERGIC53 was used as a marker of the ERGIC, and RIBOPHORIN-1 as a marker of the ER. SEC23B bands were subjected to densitometry analysis and quantified as percentage of total SEC23B or SEC23B(S186D). (B) Schematic representation of the SEC23B genomic locus and gRNA target location. Exon five refers to human SEC23B gene (Gene ID: 10483). Wild-type genomic DNA template and knock-in mutant sequences are shown for both SEC23B(S186A) and SEC23B(S186N).
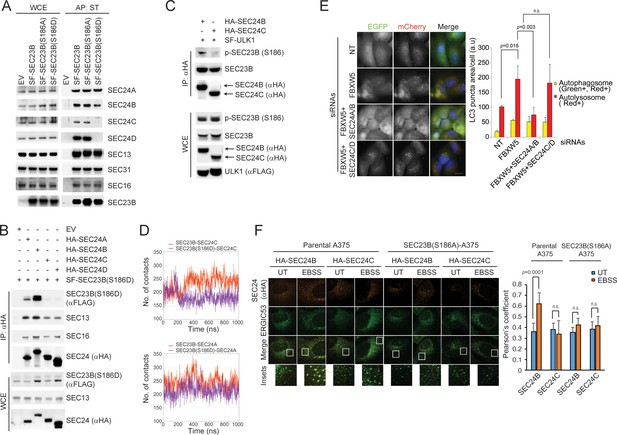
SEC24A/B, but not SEC24C/D, specifically associate with phosphorylated SEC23B and contribute to autophagy.
(A) HEK293T cells were transfected with an EV, SF-SEC23B, or SF-SEC23B mutants. Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for affinity-purification (AP) with Streptactin (ST) beads and immunoblotting. (B) HEK293T cells were transfected with an EV or SF) SEC23B(S186D) in combination with either HA-tagged SEC24 paralogs as indicated. Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for immunoprecipitation (IP) with anti-HA beads and immunoblotting. (C) HEK293T cells were transfected with either HA-tagged SEC24B or HA-tagged SEC24C together with SF-ULK1. Twenty-four hours after transfection, cells were treated with MLN4924 for 4 hr before harvesting them for immunoprecipitation (IP) with anti-HA beads and immunoblotting. (D) Evolution of the inter-molecular contacts between monomers in the four studied systems during 1 μs molecular dynamics simulations. Upper panel: SEC23B-SEC24C (red), SEC23B(S186D)-SEC24C (violet); bottom panel: SEC23B-SEC24A (red), SEC23B(S186D)-SEC24A (violet). Contacts were calculated as the number of heavy atom interacting pairs within a distance of 4.4 Å. (E) U-2OS cells stably expressing tandem fluorescent-tagged LC3 (pBabe-mCherry-EGFP-LC3) were transfected with a NT oligo or a FBXW5 siRNA oligo in combination with the indicated siRNA oligos. Forty-eight hours after transfection, cells were replated onto coverglass followed by fixation twenty-four hours after replating. Images of mCherry-EGFP-LC3 puncta underwent automated processing with at least 100 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm (F) A375 parental cells or SEC23B(S186D)-A357 knock-in cells were transfected with either HA-tagged SEC24B or HA-tagged SEC24C. Twenty-four hours after transfection, cells were either left untreated (UT) or starved with EBSS for two hours. Next, cells were fixed for immunofluorescence as indicated. Images were analysed by ImageJ with at least 100 cells counted per sample. Quantification of SEC24 overlapped with ERGIC53 was performed using the Pearson's correlation coefficient. The data are presented as mean ±SD (right panel). Scale bar, 10 μM.
-
Figure 6—source data 1
Source data for Figure 6E and F.
- https://doi.org/10.7554/eLife.42253.019
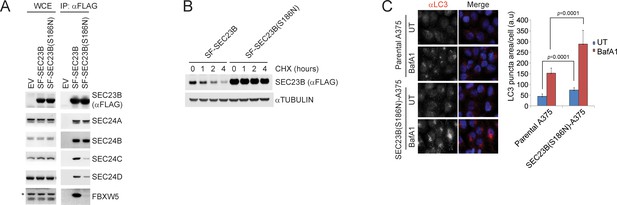
A human melanoma-associated mutation in SEC23B results in its stabilization and increased autophagy flux.
(A) HEK239T cells were transfected with either SF-SEC23B or SF-SEC23B(S186N). Twenty-four hours after transfection, cells were harvested for immunoprecipitation (IP) with FLAG-M2 beads and immunoblotted as indicated. (B) HEK293T cells were transfected with either Streptag-FLAG-tagged (SF) wild-type SEC23B (SF-SEC23B) or Streptag-FLAG-tagged (SF) SEC23B(S186N). Twenty-four hours after transfection, cells were treated with cycloheximide (CHX) for the indicated time and subjected to immunoblot analysis. (C) A375 parental cells or SEC23B(S186N)-A357 knock-in cells were treated with BafA1 and fixed for immunofluorescence. Images of endogenous LC3 puncta underwent automated processing with at least 100 cells counted per sample. The data are presented as mean ±SD (right panel). Scale bar, 10 μm.
-
Figure 7—source data 1
Source data for Figure 7C.
- https://doi.org/10.7554/eLife.42253.021
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | FBXW5 | This paper | Yenzyme | (1:250 dilution for western blotting) |
| Antibody | Phospho- SEC23B (S186) | This paper | Yenzyme | (1:200 dilution for western blotting) |
| Antibody | SEC23A | Randy Schekman’s lab | PMID: 24069399 | (1:1000 dilution for western blotting) |
| Antibody | SEC23B | Randy Schekman’s lab | PMID: 24069399 | (1:1000 dilution for western blotting) |
| Antibody | SEC24A | Randy Schekman’s lab | PMID: 18056412 | (1:2000 dilution for western blotting) |
| Antibody | SEC24B | Randy Schekman’s lab | PMID: 18056412 | (1:2000 dilution for western blotting) |
| Antibody | SEC24C | Randy Schekman’s lab | PMID: 18056412 | (1:2000 dilution for western blotting) |
| Antibody | SEC24D | Randy Schekman’s lab | PMID: 18056412 | (1:2000 dilution for western blotting) |
| Antibody | FLAG | Sigma-Aldrich | Cat# F7425 | (1:1000 dilution for western blotting, 1:1000 for immunofluorescence) |
| Antibody | HA | Covance | Cat# MMS- 101P | (1:1000 dilution for western blotting) |
| Antibody | MYC | Sigma-Aldrich | Cat# M5546 | (1:1000 dilution for western blotting) |
| Antibody | SAR1 | Thermo Scientific | Cat# PA5 -27642 | (1:5000 dilution for western blotting) |
| Antibody | CUL1 | Life Technologies | Cat# 322400 | (1:1000 dilution for western blotting) |
| Antibody | SKP1 | Michele Pagano’s lab | Yenzyme | (1:1000 dilution for western blotting) |
| Antibody | SEC13 | Bethyl Laboratories | Cat# A303 -980A | (1:10,000 dilution for western blotting) |
| Antibody | SEC31A | Bethyl Laboratories | Cat# A302 -336A | (1:10,000 dilution for western blotting) |
| Antibody | ULK1 | Cell Signaling Technology | Cat# 8054 s | (1:1000 dilution for western blotting) |
| Antibody | Phospho-ULK1 (S757) | Cell Signaling Technology | Cat# 6888 s | (1:1000 dilution for western blotting) |
| Antibody | LC3B | Novus Biologicals | Cat# NB100 -2220 | (1:10,000 dilution for western blotting, 1:1000 for immunofluorescence) |
| Antibody | Phospho- ATG13 (S318) | Rockland Immunochemicals | Cat# 600– 401 C49S | (1:1000 dilution for western blotting) |
| Antibody | Phospho- Beclin-1 (S15) | Abbiotec | Cat# 254515 | (1:500 dilution for western blotting) |
| Antibody | Beclin-1 | Santa Cruz Biotechnology | Cat# SC-48381 | (1:1000 dilution for western blotting) |
| Antibody | GAPDH | Cell Signaling Technology | Cat# 97166S | (1:10,000 dilution for western blotting) |
| Antibody | ACTIN | Sigma-Aldrich | Cat# A5441 | (1:10,000 dilution for western blotting) |
| Antibody | Tubulin | Sigma-Aldrich | Cat# T6074 -200UL | (1:10,000 dilution for western blotting) |
| Antibody | ERGIC53 | Sigma-Aldrich | Cat# E1031 -200UL | (1:5000 dilution for western blotting, 1:1000 for immunofluorescence) |
| Antibody | SEC16 | Bethyl Laboratories | Cat# A300-648A | (1:5000 dilution for western blotting) |
| Antibody | FBXO11 | Novus Biologicals | Cat# H00080204 -B01 | (1:1000 dilution for western blotting) |
| Antibody | FBXW9 | Michele Pagano’s lab | (1:1000 dilution for western blotting) | |
| Peptide, recombinant protein | MG132 | Peptide international | Cat# IZL-3175v | (final concentration, 5 mM) |
| Chemical compound, drug | MLN4924 | Active Biochem | Cat# A-1139 | (final concentration, 1 mM) |
| Chemical compound, drug | Cycloheximide | Sigma-Aldrich | Cat# C7698-1G | (final concentration, 50 ng/ml) |
| Chemical compound, drug | Bafilomycin A1 | Santa Cruz Biotechnology | Cat# sc-201550A | (final concentration, 0.1 mg/ml) |
| Chemical compound, drug | Polybrene | Sigma-Aldrich | Cat# H9268-10G | (final concentration, 8 μg/ml) |
| Chemical compound, drug | SBI-0206965 | Selleck Chemicals | Cat# S7885 | (final concentration, 1 mM) |
| Peptide, recombinant protein | SEC23B peptide | This paper | Yenzyme | |
| Peptide, recombinant protein | SEC23B phospho- peptide | This paper | Yenzyme | |
| Cell line (human) | HEK293T | ATCC | Cat# CRL-3216 | |
| Cell line (human) | U-2OS | ATCC | Cat# HTB-96 | |
| Cell line (human) | RPE1-hTERT | ATCC | Cat# CRL-4000 | |
| Cell line (human) | A375 | ATCC | Cat# CRL-1619 | |
| Cell line (human) | HeLa | ATCC | Cat# CCL-2 | |
| Commercial assay or kit | Pierce Gaussia Luciferase Flash Assay Kit | Thermo Scientific | Cat# 16159 | |
| Other | siRNAs to FBXW5 (#1) | Dharmacon | Cat# J-013389 -05-0002 | Oligonucleotides |
| Other | siRNAs to FBXW5 (#2) | Dharmacon | Cat# J-013389 -06-0002 | Oligonucleotides |
| Other | siRNAs to FBXW5 (#3) | Dharmacon | Cat# J-013389 -07-0002 | Oligonucleotides |
| Other | siRNAs to FBXW5 (#4) | Dharmacon | Cat# J-013389 -08-0002 | Oligonucleotides |
| Other | siRNAs to FBXW5 | Santa Cruz Biotechnology | Cat# sc-92629 | Oligonucleotides |
| Other | siRNAs to SEC23A | Dharmacon | Cat# M-009582 -00-0005 | Oligonucleotides |
| Other | siRNAs to SEC23B | Dharmacon | Cat# M-009592 -01-0005 | Oligonucleotides |
| Other | siRNAs to ULK1 | Santa Cruz Biotechnology | Cat# sc-44182 | Oligonucleotides |
| Other | siRNAs to ULK2 | Santa Cruz Biotechnology | Cat# sc-44183 | Oligonucleotides |
| Other | siRNAs to SEC24A | Dharmacon | Cat# L-024405 -01-0005 | Oligonucleotides |
| Other | siRNAs to SEC24B | Dharmacon | Cat# L-008299 -02-0005 | Oligonucleotides |
| Other | siRNAs to SEC24C | Dharmacon | Cat# L-008467 -02-0005 | Oligonucleotides |
| Other | siRNAs to SEC24D | Dharmacon | Cat# L-008493 -01-0005 | Oligonucleotides |
| Other | Non-targeting siRNA (CGUACGCGGAAUACUUCGA) | Dharmacon | Oligonucleotides | |
| Recombinant DNA reagent | pCS2 + 3x HA-Sec24A | Randy Schekman’s lab | ||
| Recombinant DNA reagent | pCS2 + 3x HA-Sec24B | Randy Schekman’s lab | ||
| Recombinant DNA reagent | pCS2 + 3x HA-Sec24C | Randy Schekman’s lab | ||
| Recombinant DNA reagent | pCS2 + 3x HA-Sec24D | Randy Schekman’s lab | ||
| Recombinant DNA reagent | pBabe-puro- mCherry-EGFP-LC3B | Addgene | Cat# 22418 | |
| Recombinant DNA reagent | pcdna6.2- myc ULK1 wt | Addgene | Cat# 27629 | |
| Recombinant DNA reagent | pcdna6.2- myc ULK1 k46I | Addgene | Cat# 27630 | |
| Recombinant DNA reagent | GFP-SAR1 (T39N) | Antonella De Matteis’ lab | ||
| Recombinant DNA reagent | GFP-SAR1 (H79G) | Antonella De Matteis’ lab |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42253.022





