Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response
Figures
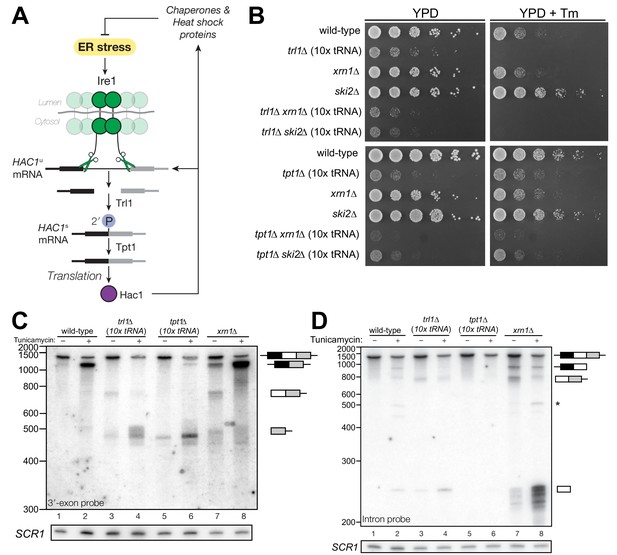
HAC1 mRNA processing defects in RNA repair and decay mutants.
(A) Schematic of the budding yeast unfolded protein response. ER stress activates Ire1 (green), which excises an intron (thin line) from HAC1u mRNA. The 5′ (black) and 3′ (grey) exons are ligated by Trl1 yielding spliced HAC1 (HAC1s) with a 2′-phosphate at the newly-formed ligation junction, which is subsequently removed by the 2′-phosphotransferase Tpt1. HAC1s mRNA is translated into a transcription factor (Hac1, purple) that upregulates the HAC1 gene itself (a positive feedback loop), as well as several chaperones and heat shock proteins that resolve the stress. (B) Yeast cells with mutations in RNA repair and decay factors were serially diluted (5-fold) and spotted onto agar media (YPD and YPD containing tunicamycin (Tm; 0.16 µg/mL)), grown at 30°C for 2 days, and photographed. The ‘10x tRNA’ plasmid encodes 10 intronless tRNAs that bypass the lethality of trl1∆ and tpt1∆ (Cherry et al., 2018). The top panels depict cells with deletions of the RNA ligase TRL1 and the bottom panels depict growth of cells deletions of the 2′-phosphotransferase TPT1. (C) HAC1 processing in RNA repair and decay mutants (3′-exon probe). HAC1u cleavage and ligation were analyzed in mutants of TRL1 RNA ligase and TPT1 2′-phosphotransferase by denaturing acrylamide gel northern blotting using a probe to the HAC1 3′-exon. Diagrams of HAC1u, HAC1s, and HAC1 splicing intermediates are drawn next to predominant bands (see Table 1 for descriptions and sizes of all annotations). HAC1u is cleaved and ligated to produce HAC1s in wild-type cells (lanes 1 and 2). Intron/3′-exon and 3′-exon splicing intermediates accumulate in trl1∆ cells, but HAC1s is not produced (lanes 3 and 4). In tpt1∆ cells, a small amount of cleaved 3′-exon is present in the absence of tunicamycin (lane 5), whereas HAC1s and 3′-exon accumulate upon tunicamycin induction (lane 6). Cells lacking xrn1∆ grown in the absence of tunicamycin produce HAC1s and Intron/3′-exon and 3′-exon splicing intermediates (lane 7), and tunicamycin addition causes an increase in production of HAC1s (lane 8). The blot was stripped and reprobed using a probe for SCR1 as a loading control. (D) HAC1 processing in RNA repair and decay mutants (intron probe). Linear intron (252 nt) is excised from HAC1u upon tunicamycin treatment (lanes 1 and 2) and linear intron is excised and accumulates in trl1∆ cells in the presence and absence of treatment (lanes 3 and 4). Excised intron is present a low levels in tpt1∆ cells (lanes 5 and 6), whereas xrn1∆ cells accumulate high levels of full-length intron and shorter, intron-derived decay intermediates (lanes 7 and 8). A star denotes excised and circularized intron, which migrates at ~500 nt.
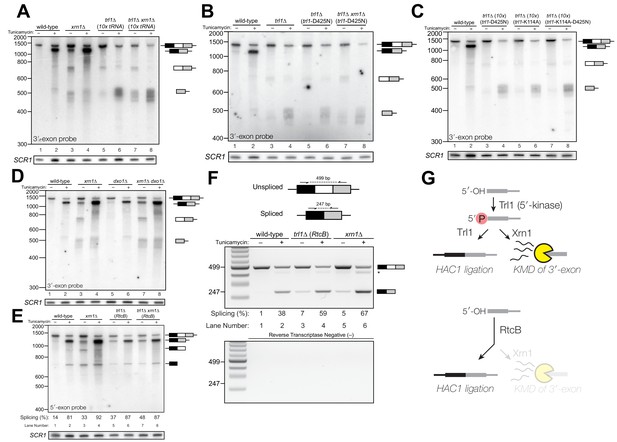
Kinase-mediated decay of HAC1 3′-exon competes with its ligation.
(A) Decay of cleaved HAC1 3′-exon requires Trl1. A northern blot for HAC1 3′-exon reveals that xrn1∆ (lanes 3 and 4) and trl1∆ (lanes 5 and 6) are mutations sufficient to cause 3′-exon to accumulate compared to wild-type. In xrn1∆ cells, the accumulation appears tunicamycin-treatment independent, whereas in trl1∆ cells the accumulation increases upon treatment. The accumulation is also present in xrn1∆ trl1∆ cells (lanes 7 and 8). (B) Decay of cleaved HAC1 3′-exon requires the catalytic activity of Trl1 5′-kinase. We expressed a kinase-inactive missense mutant of Trl1 (Wang et al., 2006), trl1-D425N, to assess the contribution of RNA 5′-kinase activity to decay of HAC1 intermediates. Expression of trl1-D425N (lanes 5 and 6) caused similar accumulation of 3′-exon as in the trl1∆ deletion, and xrn1∆ trl1-D425N cells have levels of 3′-exon similar to trl1-D425N alone (compare lanes 6 and 8), indicating that Trl1 5′-kinase activity is required for Xrn1-mediated decay. (C) 5′-kinase and ligase domains contribute to the abundance of liberated 3′-exon. We expressed a ligase-inactive missense mutant of Trl1 (Sawaya et al., 2003), trl1-K114A, to assess the contribution of ligation activity to levels of HAC1 intermediates. Expression of trl1-K114A (lanes 5 and 6) lead to a moderate accumulation of 3′-exon. The double missense mutant, trl1-K114A-D425N (lanes 7 and 8), has 3′-exon accumulation much like that of trl1-D425N, albeit stronger. (D) Dxo1 activity does not affect HAC1 3′-exon abundance or cause promiscuous ligation. Northern blot analysis of 3′-exon shows that dxo1∆ cells phenocopy wild-type cells, whereas xrn1∆ and dxo1∆ xrn1∆ cells accumulate similar levels of 3′-exon, indicating that Dxo1 does not contribute substantially to 3′-exon abundance. (E) Cells lacking Trl1 and expressing E. coli RtcB promiscuously splice HAC1s. Northern blot analysis for HAC1 5′-exon shows that trl1∆ (RtcB) cells promiscuously splice HAC1s, similar to xrn1∆ cells (compare lanes 5 and 7 to lane 3), and the defects in HAC1 splicing in trl1∆ (RtcB) cells are unaffected by xrn1∆ (compare lanes 7 and 8 to 5 and 6). (F) RT-PCR assay to measure HAC1 splicing shows promiscuous splicing of HAC1s in RtcB and xrn1∆ cells. Similar to E), here an endpoint RT-PCR assay using primers that flank the intron or splice junction assesses and semi-quantifies HAC1 splicing. Tunicamycin induces HAC1s production in wild-type cells, (lanes 1 and 2) but HAC1s is detected in in trl1∆ (RtcB) and xrn1∆ cells under normal growth conditions (without tunicamycin, lanes 3 and 5). An asterisk marks an unidentified PCR product. (G) Model for kinase-mediated decay of cleaved HAC1 3′-exon. Ire1 cleavage produces a 3′-exon with a 5′-OH that is phosphorylated by Trl1 5′-kinase. The 5′-phosphorylated product is then adenylylated and ligated to the HAC1 5′-exon by Trl1, or degraded by the 5′-phosphate-dependent 5′→3′ exonuclease Xrn1. In xrn1∆ cells (top), the lack of robust 5′→3′ decay favors ligation, leading to promiscuous splicing under normal growth conditions. In trl1∆ cells expressing RtcB (bottom), RtcB directly ligates the 5′-OH products of Ire1 cleavage, and the lack of Trl1 5′-kinase activity renders Xrn1 decay irrelevant, causing promiscuous production of HAC1s.
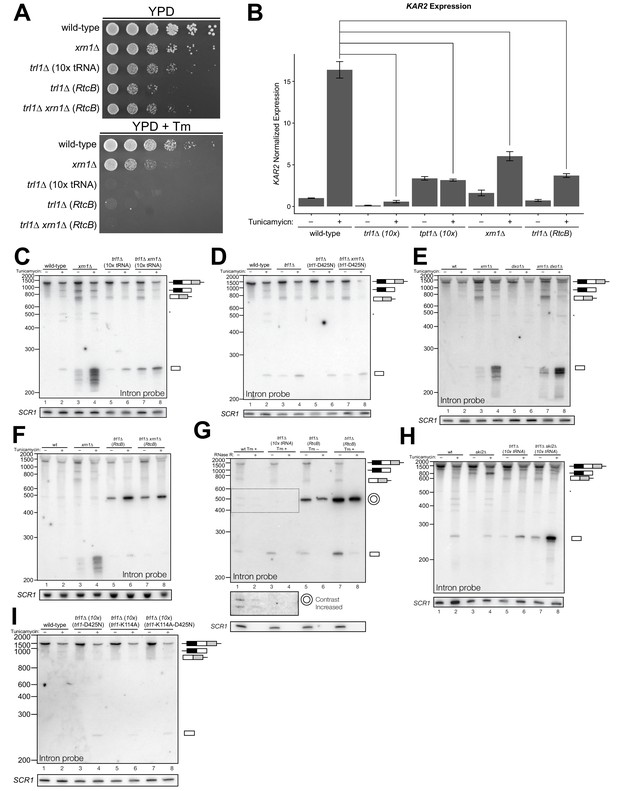
Kinase-mediated decay of excised HAC1 intron is required to activate the unfolded protein response.
(A) A serial dilution (5-fold) yeast growth assay on rich media (YPD) and tunicamycin-containing media (+Tm) compares the growth of xrn1∆, trl1∆ (10x tRNA), and trl1∆ (RtcB) cells to resist protein-folding stress. Growth of wild-type cells is modestly affected by tunicamycin, whereas growth of xrn1∆ cells is partially inhibited by tunicamycin. Cells that lack ligase (trl1∆), and cells expressing E. coli RtcB RNA ligase in lieu of TRL1 (Tanaka et al., 2011) both fail to grow on media containing tunicamycin, and this growth defect is not affected by xrn1∆. (B) Expression of a UPR-responsive gene is compromised in RNA repair and decay mutants. RT-qPCR of mRNA for KAR2 (BiP), a direct target of Hac1 (Kohno et al., 1993), performed on total RNA from the indicated genotypes shows that wild-type cells induce KAR2 expression by 16-fold upon tunicamycin treatment. (Error bars are 95% confidence interval, n = 3; comparison bars represent p<0.01, Student’s t-test.) trl1∆ cells show an insignificant increase in UPR induction, whereas tpt1∆ cells have elevated KAR2 levels in the absence of tunicamycin, which does not change significantly after tunicamycin treatment. trl1∆ (RtcB) and xrn1∆ cells have a modest increase in expression, but not to the same degree as wild-type (p<0.01). (C) Excised HAC1 intron is stabilized in xrn1∆ and trl1∆ cells. Northern blot analysis using a probe to HAC1 intron reveals that excised intron (252 nt) and partially-degraded intron intermediates accumulate in xrn1∆ cells. Ligase-delete cells (trl1∆) also accumulate intron as a uniformly sized 252 nt product. In xrn1∆ and trl1∆ cells, intron accumulates in the absence tunicamycin. (D) Catalytic activity of Trl1 5′-kinase is required for 5′→3′ decay of excised HAC1 intron. Northern blot analysis using a probe to HAC1 intron shows that a missense mutation in the 5′-kinase domain of Trl1 (trl1-D425N) phenocopies the HAC1 accumulation of trl1∆ cells (lanes 4 and 6, also C). (E) The distributive 5′→3′ exonuclease Dxo1 can partially degrade HAC1 intron. Northern blot analysis for HAC1 intron on total RNA from dxo1∆ and dxo1∆ xrn1∆ cells shows that Dxo1 can partially degrade HAC1 intron when it accumulates in xrn1∆ cells (compare lanes 4, 6 and 8). (F) A slow-migrating intron species accumulates in trl1∆ cells expressing RtcB. Northern blot analysis of RNA from wild-type and trl1∆ cells shows that wild-type cells accumulate linear, partially degraded intron (lanes 1–4), whereas trl1∆ (RtcB), and trl1∆ xrn1∆ (RtcB) cells accumulate a slower-migrating species (~500 nt; lanes 5–8). (G) Cells expressing RtcB accumulate circular HAC1 intron. To test whether the slower-migrating band was circularized, total RNA was treated with RNase R and analyzed by northern blot. The slower-migrating species is largely protected from degradation, indicating it is a circle. Linear HAC1 intron and SCR1 (bottom) are degraded upon RNase R treatment (lanes 2, 4, 6 and 8). A panel of enhanced contrast shows that the slower migrating species in wild-type cells are circular, excised HAC1 introns, resistant to RNase R. Circular intron only occurs in samples from cells expressing a ligase. (H) The cytoplasmic exosome degrades HAC1 intron when 5′→3′ decay is disabled. Northern blot analysis using a HAC1 intron probe on total RNA from ski2∆ (a component of the cytoplasmic exosome) cells showed that ski2∆ cells accumulate excised HAC1 intron (lane 4) at levels similar to wild-type (lane 3). In trl1∆ ski2∆ cells, a lack of both kinase-mediated decay (trl1∆) and 3′→5′ decay (ski2∆) causes accumulation of excised intron relative to trl1∆ cells (compare lanes 6 and 8). (I) Catalytic activity of Trl1 ligase domain contributes to processing of excised HAC1 intron. We expressed a ligase-inactive Trl1 allele, trl1-K114A, as in Figure 2C. Expression of trl1-K114A (lanes 5 and 6) lead to a modest accumulation of intron, though not to the same extent as in the kinase-inactivated mutant (trl1-D425N) (lanes 3 and 4). The double missense mutant, trl1-K114A-D425N (lanes 7 and 8), exhibits intron accumulation similar that of trl1-D425N.
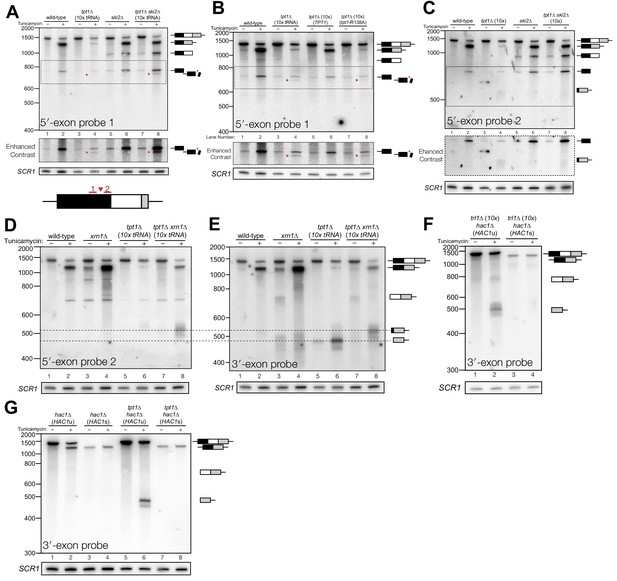
Incompletely processed HAC1s mRNA is cleaved and degraded.
(A) A shortened form of 5′-exon accumulates in tpt1∆ cells and is degraded by the cytoplasmic exosome. Northern blot analysis of tpt1∆ and tpt1∆ ski2∆ using a probe to 5′-exon identified a shortened form of liberated 5′-exon, about 50 nt smaller than full-length (red arrowheads). A region of enhanced contrast shows the specific accumulation of this product in tpt1∆ and tpt1∆ ski2∆ cells. The bottom diagram depicts the relative positions of probes 1 and 2 on the HAC1 5′-exon; a red arrowhead marks the putative site of cleavage. (B) Production of shortened 5′-exon requires the catalytic activity of Tpt1. Northern blot analysis of tpt1∆ cells expressing either a plasmid-encoded wild-type copy of TPT1 or a catalytically-inactive missense mutant (tpt1-R138A) (Sawaya et al., 2005) using a probe for 5′-exon. Production of the cleaved 5′-exon seen in tpt1∆ cells (lane 4, red arrow marks the band) is rescued by plasmid-mediated expression of wild-type TPT1 (lane 6) but not by Tpt1-R138A (lane 8, red arrow marks the band), confirming that Tpt1 catalytic activity is required to prevent HAC1s secondary cleavage. (C) The shortened 5′-exon is missing a portion of its 3′-end. A northern blot was probed with 5′-exon probe 2, which hybridizes to the 3′-most 30 nt of HAC1 5′-exon (see diagram in A). Compared to the blot in (A), the shortened 5′-exon species is absent (lanes 4 and 8) and instead the probe detects smaller bands consistent with the length of the elongated 3′-exon (see region of enhanced contrast). (D) A lengthened form of 3′-exon accumulates in tpt1∆ cells. Total RNA from tpt1∆ and xrn1∆ cells was analyzed by northern blot with 5′-exon probe 2, a probe that anneals to the 3′-most 30 nt of HAC1 5′-exon. (D) and E) share the same band interpretation key, so dashed lines have been drawn across to illustrate the different positions of typical 3′-exon (474 nt) and elongated 3′-exon (524 nt). (E) The elongated 3′-exon from tpt1∆ cells co-migrates and co-hybridizes with 5′-exon probe 2 (see D). Stripping the blot from D) and re-hybridizing it with HAC1 3′-exon probe detects the same, elongated band(s) as in the 5′-exon probe two blot (lane 8), as well as HAC1 3′-exon bands of typical length. (F) Expression of ‘pre-spliced’ HAC1s is not sufficient to promote cleavage. Total RNA from trl1∆ hac1∆ cells expressing HAC1u and HAC1s from a plasmid was analyzed by northern blot with a 3′-exon probe. Full length HAC1u expressed from this construct is cleaved upon tunicamycin addition (lane 2). Expression of ‘pre-spliced’ HAC1s yields a single product with no decay intermediates (lanes 3 and 4), indicating that ‘pre-spliced’ HAC1s—which was not produced by ligation and therefore lacks a 2′-phosphate—is not sufficient to recapitulate the cleavage found in tpt1∆ cells. (G) Expression of ‘pre-spliced’ HAC1s is not sufficient to promote secondary cleavage. Total RNA from hac1∆ and tpt1∆ hac1∆ cells expressing HAC1u and ‘pre-spliced’ HAC1s from a plasmid was analyzed by northern blot with a 3′-exon probe. In hac1∆ cells, full length HAC1u expressed from this construct is cleaved and ligated upon tunicamycin addition (lane 2). Expression of ‘pre-spliced’ HAC1s yields a single product with no processing intermediates (lanes 3 and 4), indicating that ‘pre-spliced’ HAC1s is not further processed. The HAC1u construct expressed in cells of genotype tpt1∆ hac1∆ (10x tRNA) gets cleaved upon tunicamycin treatment (lane 6), yielding a secondary cleavage fragment. Expression of pre-spliced HAC1s in tpt1∆ hac1∆ cells does not produce any additional processing fragments, indicating that 2′-phosphorylated products from HAC1u are required for secondary cleavage in tpt1∆ cells.
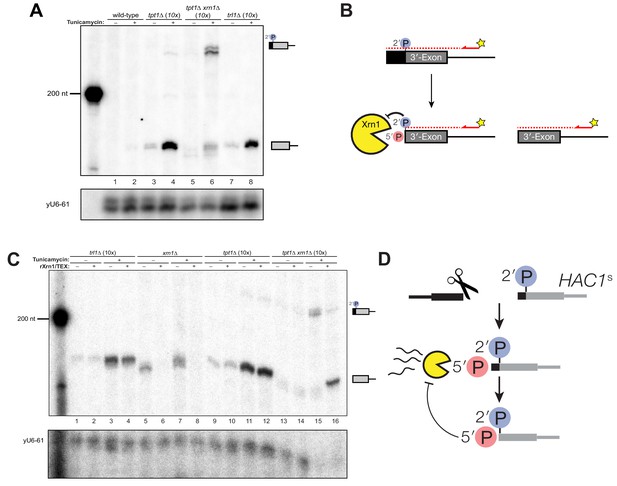
A 5′- and 2′-phosphorylated HAC1 decay intermediate inhibits Xrn1.
(A) Primer extension analysis of HAC1 3′-exon cleavage products. Primer extension using a probe for 3′-exon was performed on total RNA from wild-type, tpt1∆ (10x tRNA), tpt1∆ xrn1∆ (10x tRNA), and trl1∆ (10x tRNA) cells treated with or without tunicamycin. A loading control for U6 snRNA is depicted in the bottom panel. The extension product of cleaved 3′-splice site is 174 nt, found in wild-type cells treated with tunicamycin (lane 2) and is present in untreated trl1∆ cells but increases upon tunicamycin treatment (lanes 7 and 8). An extension product from tpt1∆ cells accumulates upon tunicamycin treatment (lanes 3 and 4) and co-migrates with the product from wild-type (lane 2) and trl1∆ (lane 8) cells. In addition, a faint elongated product at ~225 nt is present in tpt1∆ cells (lane 4). This elongated product accumulates to higher levels in tpt1∆ xrn1∆ cells treated with tunicamycin (lane 6). (B) Model of HAC1 3′-exon primer extension product lengths. A 5′-radiolabeled (yellow star) primer (HAC1 3′-exon probe, see Table 2) anneals to HAC1 3′-exon mRNA and primes cDNA synthesis by a reverse transcriptase. The reverse transcriptase stops synthesizing cDNA when it runs out of RNA template at the 5′-terminus of the 3′-exon. The model shows three situations as observed in (A): 1) canonical 3′-exon cleavage fragments (lower right), as observed in trl1∆ cells; 2) extended HAC1 3′-exon (above reaction arrow), as caused by secondary cleavage of HAC1s, observed in tpt1∆ xrn1∆ (10x) cells; and 3) extended HAC1 3′-exon on which Xrn1 (cellular or in vitro) initiated decay, but failed to proceed past the 2′-P (beneath arrow). (C) Susceptibility of 3′-exon cleavage products to in vitro Xrn1 degradation. Total RNA from (A) was treated with recombinant Xrn1 (rXrn1) and analyzed by primer extension for the 3′-exon. The loading control performed on U6 snRNA is depicted in the bottom panel. The 3′-exon from trl1∆ cells accumulates upon tunicamycin treatment (compare lanes 1 and 2 to 3 and 4) but the 3′-exon is resistant to rXrn1 (lanes 3 and 4) because it lacks a 5′-phosphate due to lack of Trl1 5′-kinase activity in these cells. In contrast, the 3′-exon products from xrn1∆ cells, which have Trl1 5′-kinase and thus 5′-phosphates, are degraded by rXrn1 (lanes 6 and 8). The 3′-exon product in tpt1∆ cells is resistant to rXrn1 treatment (compare lanes 9 to 10, and 11 to 12), despite the fact these cells have both Trl1 and Xrn1. The elongated 3′-exon product that accumulates in tpt1∆ xrn1∆ cells is partially degraded by rXrn1 (compare lane 15 to 16) and the decay intermediate co-migrates with cleaved 3′-exon. (D) Model depicting cleavage and decay of 2′-phosphorylated HAC1s. HAC1s is cleaved (likely by Ire1)~50 nt upstream of the 2′-phosphorylated ligation junction, creating a 3′-product with ~50 nt of sequence of the 5′-exon (black) and an internal 2′-phosphate. Xrn1 initiates decay at the 5′-terminus, degrading the 5′-exon portion (black) up to the site of 2′-phosphorylation. The product of partial decay contains a 5′- and 2′-phosphate at its first position, which inhibits further degradation by Xrn1.
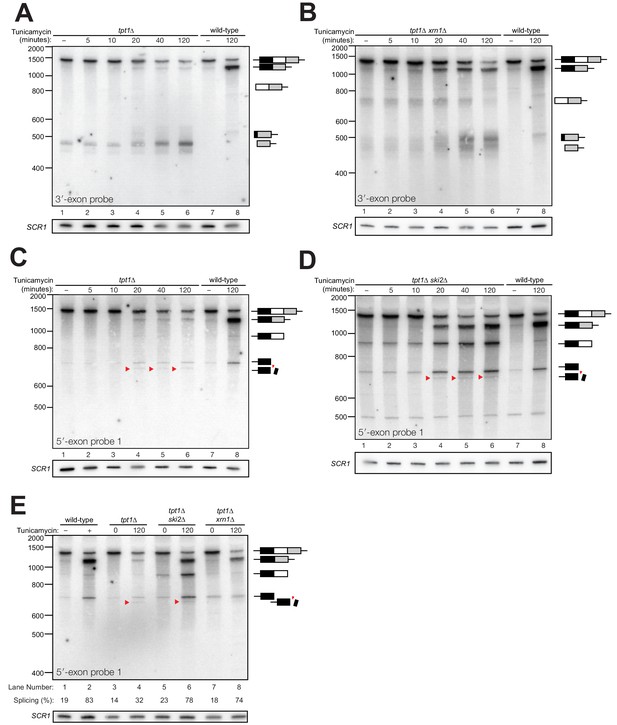
Kinetic analysis of HAC1 mRNA processing in cells lacking Tpt1.
(A) Northern blot analysis of a time course of tunicamycin treatment (0 to 120 min) in tpt1∆ (10x tRNA) cells showing the dynamics of HAC1u splicing using a probe for 3′-exon. The SCR1 loading control is shown below the panel. RNA from wild-type cells treated for 0 and 120 min tunicamycin was loaded in lanes 7 and 8. HAC1s accumulates slowly over 120 min and its abundance at the end of the time-course is 35-fold lower than wild-type (lanes 6 and 8). A 3′-exon cleavage product (~475 nt) is present at time 0 and accumulates over time; its increase is coincident with the appearance of HAC1s (20 min time point) and its abundance at 120 min exceeds the level of HAC1s. A 3′-exon cleavage product in wild-type cells (~500 nt) is apparent at 120 min (lane 8). (B) Northern blot analysis of HAC1 splicing in tpt1∆ xrn1∆ (10x tRNA) cells using a probe for 3′-exon. Conditions are the same as A). HAC1s increases at 20 min and its final level (lane 6) is higher than HAC1s in tpt1∆ cells (A), lane 6), approaching wild-type levels (lane 8). Levels of the product of 5′-splice site cleavage (intron/3′-exon) accumulate over 120 min. The 3′-exon product (~475 nt) and cleaved 3′-exon (~500 nt) begin accumulating at 20 min, coincident with increased levels of HAC1s. A 3′-exon cleavage product in wild-type cells (~500 nt) is apparent at 120 min (lane 8). (C) Northern blot analysis of HAC1 splicing in tpt1∆ (10x tRNA) cells using a probe for 5′-exon. Conditions are the same as A). HAC1s increases at 20 min up to final level. Canonical 5′-exon (728 nt) and secondarily-cleaved 5′-exon (~675 nt; red arrowheads) begin accumulating at 20 min, coincident with increased levels of HAC1s. (D) Northern blot analysis of HAC1 splicing in tpt1∆ ski2∆ (10x tRNA) cells using a probe for 5′-exon. Deletion of SKI2 in the context of tpt1∆ increases the abundance of nearly all intermediates relative to tpt1∆ alone (A). Shortened HAC1 5′-exon is marked with red arrowheads. The product of 3′-splice site cleavage (5′-exon/intron) is present at time 0 and accumulates over 120 min, whereas HAC1s accumulates beginning at 20 min. (E) Northern blot analysis of HAC1 splicing using a probe for 5′-exon to compare the beginning and end time points of time course experiments performed on tpt1∆, tpt1∆ xrn1∆, and tpt1∆ ski2∆. Results from densitometry of the spliced and unspliced bands are written beneath the blot. Less HAC1 splicing takes place in tpt1∆ cells compared to wild type, and disabling decay factors ski2∆ and xrn1∆ in the tpt1∆ mutant lead to increased HAC1 splicing, compared to tpt1∆ alone.
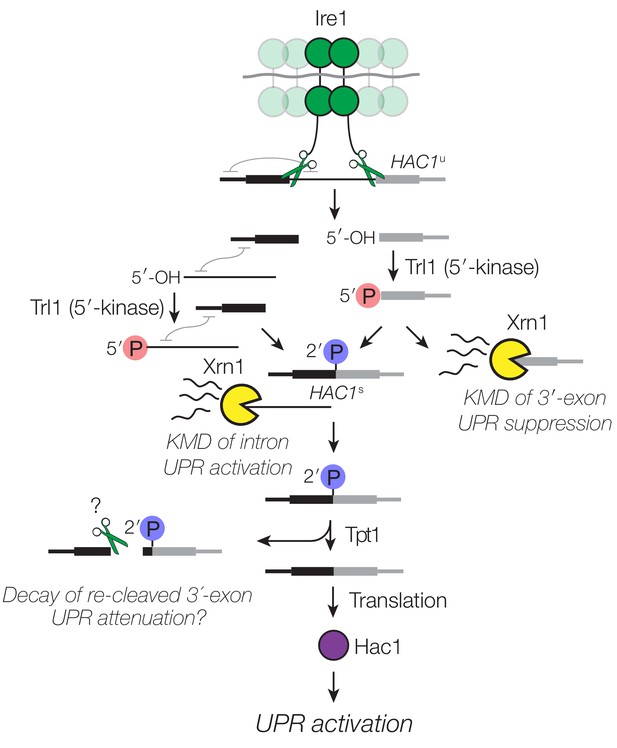
Decay of HAC1 splicing intermediates regulates UPR activation, suppression, and attenuation.
Activation of Ire1 by ER stress activates its endoribonuclease activity, leading to cleavage of the 5′- and 3′-splice sites of HAC1u. The cleaved 3′-exon (grey) is phosphorylated by the 5′-kinase activity of Trl1, permitting either its ligation or decay by Xrn1. The 5′-exon (black) and intron (thin line) form an extensive base-pairing interaction (grey squiggle). Kinase-mediated decay of the intron is required to activate the translation of HAC1s. Ligated but 2′-phosphorylated HAC1s may also be cleaved by Ire1 upstream of the ligation junction, and the cleavage products are degraded by Xrn1 and the cytoplasmic exosome. In cells lacking Tpt1, a unique KMD intermediate accumulates with 5′- and 2′-phosphates, which inhibit Xrn1-mediated degradation.
Tables
HAC1 processing intermediates.
https://doi.org/10.7554/eLife.42262.004| Name | Size (nt) | Visual summary | Description |
|---|---|---|---|
| HAC1u | 1450* |  | Full-length, genomic HAC1 transcript |
| HAC1s | 1198* |  | Spliced HAC1; intron removed |
| HAC1 5′-exon | 728 |  | Everything 5′ of the intron |
| Cleaved 5′-exon | ~678 |  | Fragment of 5′-Exon missing ~ 50 nt off its 3′-end |
| HAC1 intron | 252 |  | Liberated intron (alone) |
| circularized intron | ~500 |  | Circularized intron, visible in wild-type and RtcB cells |
| HAC1 3′-exon | 474* |  | Everything 3′ of the intron |
| Cleaved 3′-exon | ~524* |  | 3′-Exon with ~ 50 nt of 5′-Exon on its 5′-end |
| 5′-exon + intron | 980 |  | 5′-exon + Intron |
| Intron + 3′-exon | 726* |  | Intron + 3′-exon |
-
Sizes of HAC1 processing intermediates are predicted from strand-specific RNA sequencing data (Levin et al., 2010) mapped to the sacCer1 genome. *Size does not include poly(A) tail.
Oligonucleotide sequences.
https://doi.org/10.7554/eLife.42262.011| Oligonucleotide name | Oligonucleotide sequence (5′→3′) |
|---|---|
| HAC1-F RT-PCR | ACCTGCCGTAGACAACAACAAT |
| HAC1-R RT-PCR | AAAACCCACCAACAGCGATAAT |
| KAR2 qPCR F | AAGACAAGCCACCAAGGATG |
| KAR2 qPCR R | AGTGGCTTGGACTTCGAAAA |
| PGK1 qPCR F | TCTTAGGTGGTGCCAAAGGTT |
| PGK1 qPCR R | GCCTTGTCGAAGATGGAGTC |
| HAC1 5′-exon probe 1 | AAGTCTCTTGGTCCGACGCGGAATCGCGCA |
| HAC1 5′-exon probe 2 | CTGGATTACGCCAATTGTCAAGATCAATTG |
| HAC1 intron probe 1 | AACCGGCTCCTCCCCCATCAGAGAACCACGA |
| HAC1 intron probe 2 | GGACAGTACAAGCAAGCCGTCCATTTCTTAGT |
| HAC1 3′-exon probe (primer extension and northern) | ACCGGAGACAGAACAGTAGAAACCACTAAGCG |
| KAR2 probe | ACCGTAGGCAATGGCGGCTGCGGTTGGTTC |
| SCR1 probe (oRP100) | GTCTAGCCGCGAGGAAGG |
Strain numbers and genotypes.
All strains are background W303 (MATa {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15}).
| Strain ID | Genotype | Source |
|---|---|---|
| YJH682 | ‘wild-type’ (CRY1) | Mingxia Huang |
| YJH632 | xrn1∆::HygMX | |
| YJH745 | ski2∆::NatMX | |
| YJH898 | dxo1∆::KanMX | |
| YJH867 | xrn1∆::HygMX dxo1∆::KanMX | |
| YJH829 | tpt1∆::LEU2 (TPT1 CEN ARS URA3) | Schwer et al., 2004 |
| YJH830 | tpt1∆::LEU2 (TPT1 CEN ARS URA3) (pAG424-ccdB) | |
| YJH832 | tpt1∆::LEU2 (TPT1 CEN ARS URA3) (pAG424-10x-tRNA) | |
| YJH834 | tpt1∆::LEU2 (pAG424-10x-tRNA) | |
| YJH980 | tpt1∆::LEU2 (pAG424-10x-tRNA) (pAG413-NPr-TPT1) | |
| YJH891 | tpt1∆::LEU2 (pAG424-10x-tRNA) (pAG413-NPr-tpt1-R138A) | |
| YJH902 | tpt1∆::LEU2 xrn1∆::HygMX (pAG424-10x-tRNA) | |
| YJH901 | tpt1∆::LEU2 ski2∆::NatMX (pAG424-10x-tRNA) | |
| YJH681 | trl1∆::KanMX (pRS416-TRL1) | Schwer et al., 2004 |
| YJH708 | trl1∆::KanMX (pRS416-TRL1) (pAG424) | |
| YJH709 | trl1∆::KanMX (pRS416-TRL1) (pAG424-10x-tRNA) | |
| YJH835 | trl1∆::KanMX (pAG424-10x-tRNA) | |
| YJH887 | trl1∆::KanMX (pAG424-10x-tRNA) (pRS413-TRL1) | |
| YJH811 | trl1∆::KanMX (pAG424-10x-tRNA) (pRS413-trl1-D425N) | |
| YJH812 | trl1∆::KanMX xrn1∆::HygMX (pAG424-10x-tRNA) (pRS413-trl1-D425N) | |
| YJH912 | trl1∆::KanMX (pAG424-10x-tRNA) (pRS413-trl1-K114A) | |
| YJH913 | trl1∆::KanMX (pAG424-10x-tRNA) (pRS413-trl1-K114A-D425N) | |
| YJH808 | trl1∆::KanMX (pRS423-TPI-RtcB) | |
| YJH809 | trl1∆::KanMX xrn1∆::HygMX (pRS423-TPI-RtcB) | |
| YJH899 | trl1∆::KanMX xrn1∆::HygMX (pAG424-10x-tRNA) | |
| YJH900 | trl1∆::KanMX ski2∆::NatMX (pAG424-10x-tRNA) | |
| YJH903 | trl1∆::KanMX hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1u) | |
| YJH904 | trl1∆::KanMX hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1s) | |
| YJH920 | hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1u) | |
| YJH921 | hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1s) | |
| YJH923 | tpt1∆::LEU2 hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1u) | |
| YJH924 | tpt1∆::LEU2 hac1∆::NatMX (pAG424-10x-tRNA) (pAG413-GPD-HAC1s) |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42262.013





