Basalin is an evolutionarily unconstrained protein revealed via a conserved role in flagellum basal plate function
Figures
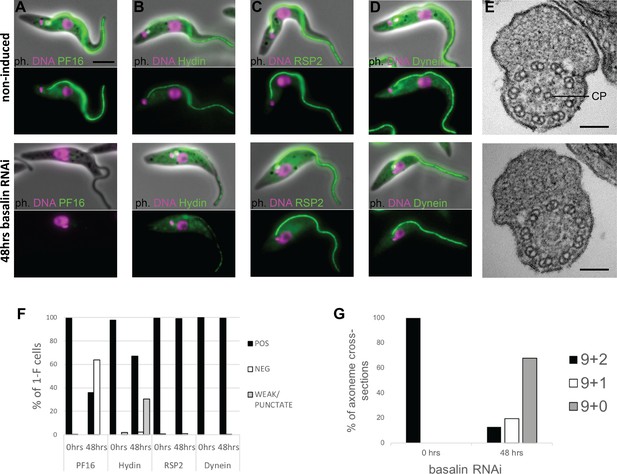
Basalin ablation causes central pair defects.
Basalin RNAi was performed in cells expressing tagged markers for different axonemal components, showed that: (A) PF16 (Tb927.1.2670, C1 projection) is absent from affected flagella, (B) hydin (Tb927.6.3150, C2 projection) is reduced and punctate, but (C) RSP2 (Tb927.5.2850, radial spokes marker) and (D) dynein (Tb927.7.820, dynein arm marker) flagellar localisation are unaffected. (E) TEM cross-sections through the axoneme demonstrate that the central pair microtubules are missing from the axonemes in cells induced for basalin RNAi. (F) The presence of axoneme marker proteins (N > 200 for each marker at each timepoint) and (G) the number of axonemal microtubules were quantified before (N = 15) and after (N = 31) basalin RNAi.
-
Figure 1—source data 1
Quantification of axonemal marker presence in 1F cells after 48 hours basalin RNAi.
- https://doi.org/10.7554/eLife.42282.005
-
Figure 1—source data 2
Quantification of axonemal microtubule arrangement after 48 hours basalin RNAi.
- https://doi.org/10.7554/eLife.42282.006
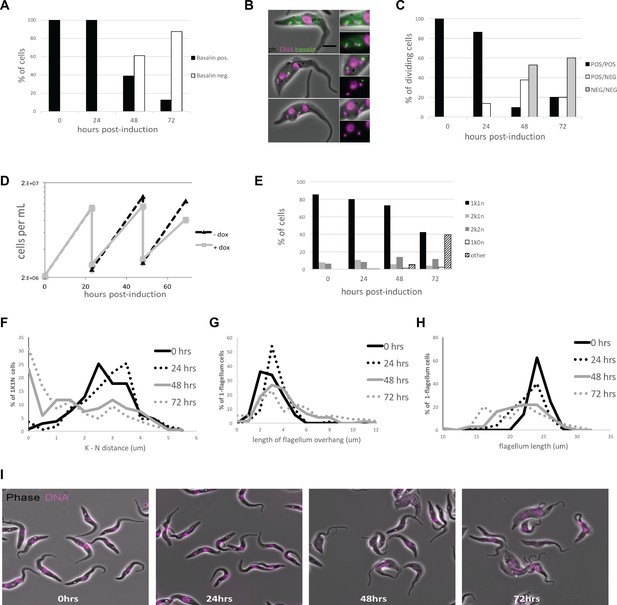
Quantitation of basalin knockdown and phenotype in trypanosome cells.
(A) Basalin was ablated in cells expressing mNeonGreen::basalin and cytoskeletons were scored on whether they were positive or negative for basalin at the TZ. RNAi caused ablation such that > 80% of cells did not have detectable basalin at 72 hours. (B) Examples of dividing cells undergoing basalin RNAi. Non-induced dividing cells have four basalin spots that corresponds to localisation at the TZ and pro-basal bodies of both old and new flagella. In cells induced for basalin RNAi where the signal is absent from new flagella, there is only one spot that is associated with the old transition zone and the pro-basal body signal is absent. (C) Quantitation of basalin signal at the old and new flagella in dividing cells shows that dividing cells with positive old/negative new (POS/NEG) are first observed after 24 hours RNAi and increase in frequency to almost 40% after 48 hours. (D) Growth rates of cells non-induced and induced for basalin RNAi demonstrate that growth rate defect is first observed between 24–48 hours, co-incident with the first cells that are negative for basalin. Representative data of three biological replicates is shown. (E) Staining the DNA of cells using Hoechst shows that only the expected kinetoplast-nucleus numbers are observed at 0 hours (i.e. 1k1n, 2k1n, 2k2n), but there is an increase in the proportion of aberrant cells after 48 and 72 hours basalin RNAi. (F) There is an accumulation of aberrant morphologies after RNAi. This includes G) a reduction in the kinetoplast-nucleus (k–n) distance, (H) an increase in the length of the flagellum that extends beyond the anterior of the cell body (flagellum overhang), and I) a reduction in the overall flagellum length.
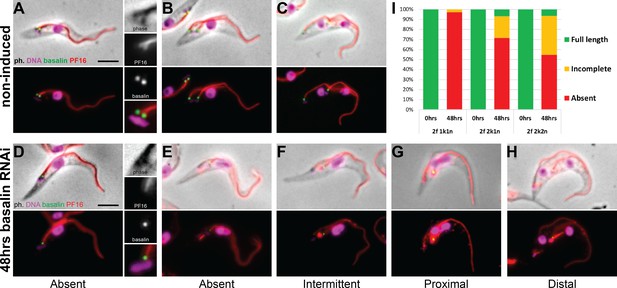
Basalin ablation inhibits central pair nucleation.
Basalin was ablated in trypanosomes expressing mNG::basalin and PF16::mScarlet-I to determine the effect of basalin ablation upon newly built axonemes. (A–C) Shows non-induced cells. (A) A cell with one nucleus and one kinetoplast (1k1n) shows PF16 incorporation into the axoneme in the early stages of flagellum construction. (B) and C) show cells with 2k1n and 2k2n, respectively. (D–H) Shows cells after 48 hours basalin RNAi induction. (D) A flagellum in the very early stages of construction is missing both basalin and PF16. Older new flagella that are absent for basalin are either D) missing PF16 altogether or have incomplete PF16 assembly into the flagellum, including F) a fragmented or intermittent signal, (G) a proximal signal that does not extend to the flagellum tip, and H) a distal signal that is missing from the proximal portion of the axoneme. (I) Quantification of non-induced vs induced dividing cells at different cell-cycle positions shows that the probability of PF16 being assembled into newly-made axonemes increases with cell-cycle progression (non-induced 1k1n N = 22, 2k1n N = 50, 2k2n N = 45; induced 1k1n N = 38, 2k1n N = 60, 2k2n N = 62).
-
Figure 2—source data 1
Quantification of axonemal PF16 incorporation in dividing cells after 48 hours basalin RNAi.
- https://doi.org/10.7554/eLife.42282.008
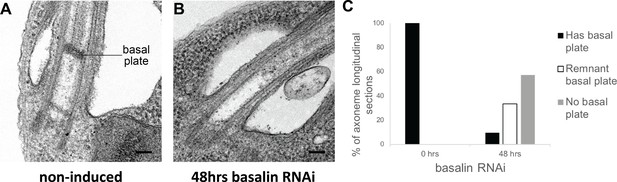
Basalin RNAi produces cells without a basal plate in the flagellum.
(A) TEM longitudinal sections through the centre of the proximal flagellum (including the transition zone and the proximal end of the axoneme), before (A) and after (B) RNAi. (C) The presence and absence of a basal plate in transition zone longitudinal sections was quantified at 0 (N = 22) and 48 hours (N = 19) basalin RNAi induction.
-
Figure 3—source data 1
Quantification of basal plate presence in TZ longitudinal sections after 48 hours basalin RNAi.
- https://doi.org/10.7554/eLife.42282.010
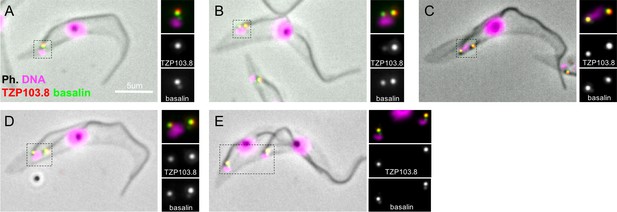
Basalin and TZP103.8 localisation through the cell cycle.
(A) In non-dividing cells, basalin localises just proximal to the TZP103.8 TZ signal and faintly to the pro-basal body. (B) As the pro-basal body matures an moves to the posterior relative to the old flagellum, the basalin signal at the new transition zone increases in intensity and a faint TZP103.8 signal is detected. (C) – (E) The TZP103.8 signal intensity increases at later stages of the cell cycle, and basalin is detected on the newly formed pro-basal bodies of both old and new flagella.
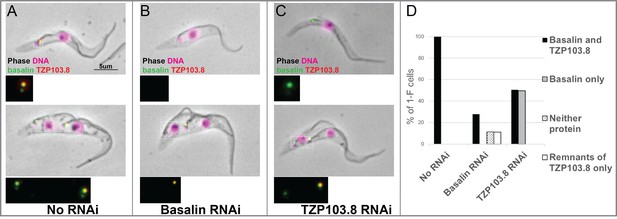
TZP103.8 depends on basalin for its localisation to the transition zone.
Basalin or TZP103.8 were ablated by RNAi in cells co-expressing mScarlet::TZP103.8 (red) and mNeonGreen::basalin (green) to determine their dependency relationship at the transition zone. (A) In uninduced cells, basalin and TZP103.8 were both detected at the transition zone. (B) When basalin was ablated by RNAi, TZP103.8 was not efficiently recruited to the transition zone. (C) In contrast, when TZP103.8 was ablated, basalin still localised to the transition zone. Top panels show cytoskeletons prepared from non-dividing cells, bottom panels show cytoskeletons prepared from dividing cells. (D) Quantification of basalin/TZP103.8 presence at the transition zone in non-induced cells (N > 150) and after 48 hours RNAi of basalin (N = 168) or TZP103.8 (N = 127) confirms the differential dependencies at the transition zone.
-
Figure 5—source data 1
Quantification of basalin and TZP103.8 presence at the TZ after 48 hours basalin or TZP103.8 RNAi.
- https://doi.org/10.7554/eLife.42282.014
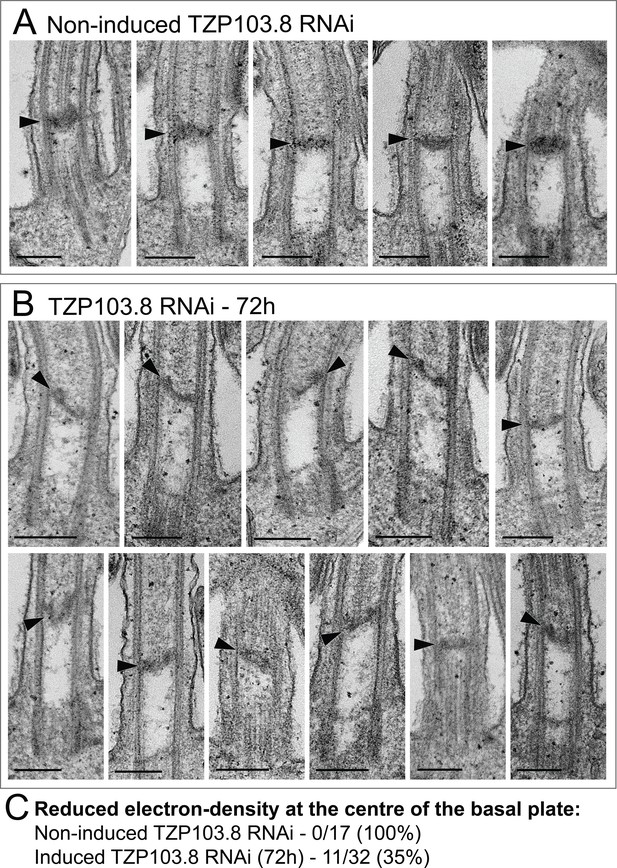
Thin-section transmission electron microscopy (TEM) analysis of the basal plate after TZP103.8 RNAi.
Longitudinal TEM sections through the proximal region of the flagellum were analyzed for the presence and morphology of the basal plate (arrowheads). (A) Representative images of showing the basal plate in non-induced cells. (B) All 11/32 longitudinal images that show a reduction in the electron density at the centre of the basal plate, after 72 hours of TZP103.8 RNAi. (C) Table showing the proportion of longitudinal images of the proximal flagellum displaying basal plate reduction, in induced and non-induced samples. Scale bars, 200 nm.
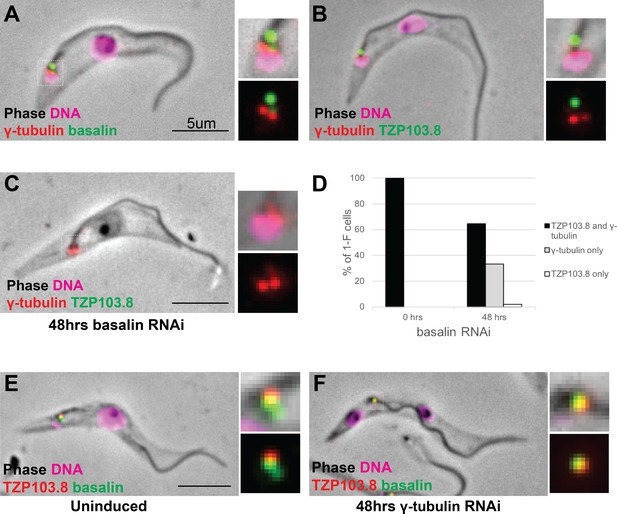
Co-localisation and dependency of gamma tubulin with basalin and TZP103.8.
Gamma tubulin localises to the base of the flagellum more proximal to (A) basalin and (B) TZP103.8. (C) and (D) Ablating basalin does not prevent gamma tubulin from being recruited to the base of the flagellum (0 hr N = 107, 48 hr N = 102). (E) and (F) Ablating gamma tubulin does not prevent basalin or TZP103.8 from being recruited to the TZ (N > 100).
-
Figure 6—source data 1
Quantification of TZP103.8 and gamma tubulin presence at the TZ in 1F cells after 48 hours basalin RNAi.
- https://doi.org/10.7554/eLife.42282.016
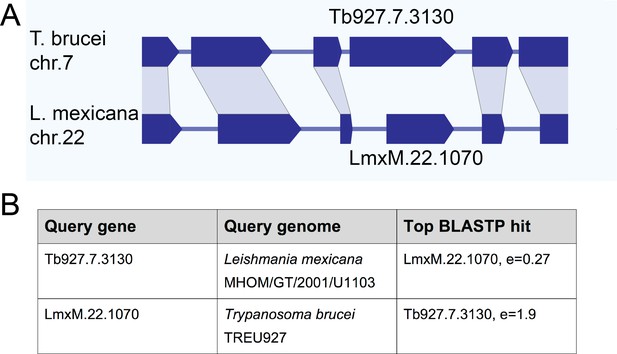
Gene synteny and sequence homology between basalin (Tb927.7.3130) and LmxM.22.1070.
(A) Basalin and LmxM.22.1070 genes are at syntenic positions along chromosome Leishmania mexicana chromosome 22 and Trypanosoma brucei chromosome 7, respectively. (B) Basalin and LmxM.22.1070 are very weak reciprocal best BLAST-P hits.

A bioinformatics comparison of basalin and LmxM.22.1070.
(A) A protein sequence alignment between basalin and its T. congolense ortholog (TcIL3000_7_2290). (B) A HHpred-driven comparison between basalin and LmxM.22.1070 demonstrates that most sequence conservation is in the helical segments. (C) Both proteins have coiled-coils towards the C terminus of the protein.

Basalin and LmxM.22.1070 are predicted to have extensive disorder.
(A) Basalin and LmxM.22.1070 show similar levels of intrinsic disorder, with ~65% disordered residues and 15–17 regions of disorder. (B) Quick2D predicts extensive disorder in basalin and LmxM.22.1070 with helices at the N and C termini (Hanson et al., 2017; Zimmermann et al., 2018; Jones and Cozzetto, 2015; Dosztányi et al., 2005).
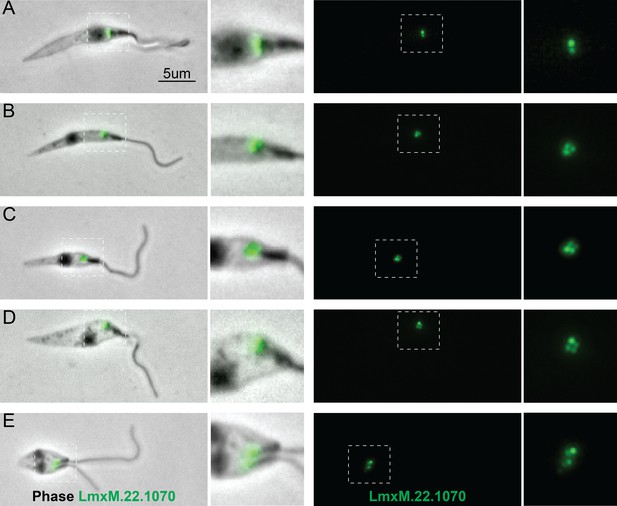
Localisation of LmxM.22.1070 in L. mexicana promastigotes through the cell cycle recapitulates that of basalin in trypanosomes.
LmxM.22.1070 was tagged at its endogenous locus with an N terminal mNeonGreen using CRISPR/CAS9 (Beneke et al., 2017) and its localisation throughout the cell cycle visualised in methanol-fixed ‘cytoskeletons’. (A) In non-dividing cells, LmxM.22.1070 localises to a position consistent with the transition zone and the pro-basal body. (B) - E) As the cell divides, the basal bodies are duplicated and LmxM.22.1070 is detected on the basal body and pro-basal body of both flagella in dividing cells.
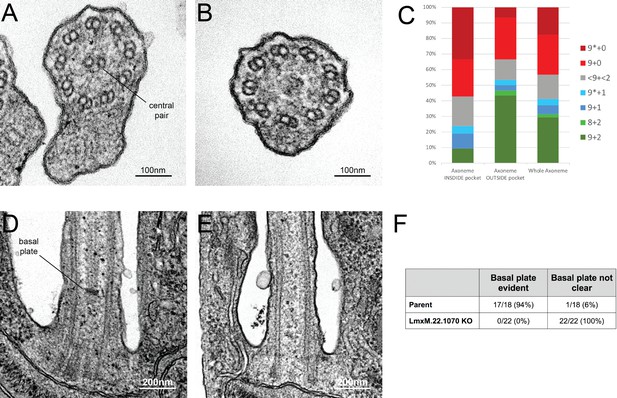
Knockout of LmxM.22.1070 in L. mexicana promastigotes causes flagella to be built without the central pair microtubules and a basal plate.
(A) and (B) TEM cross-sections through the axoneme show that the central pair is absent in knockout cells. (C) Quantification of the microtubules arrangement in KO axonemes shows that > 40% of cross sections (N = 51) are missing their CP, and that this phenotype is more penetrant in the proximal axoneme (N = 21) versus the distal axoneme (N = 30). The asterisk refers to the microtubules being disorganised. (D) and (E) TEM longitudinal sections through the transition zone demonstrate that the basal plate is absent. (F) Quantification shows that the basal plate was not observed any the transition zones that were examined in KO cells.
-
Figure 9—source data 1
Quantification of axoneme microtubule arrangement in Leishmania mexicana LmxM.22.1070 KO cells.
- https://doi.org/10.7554/eLife.42282.024
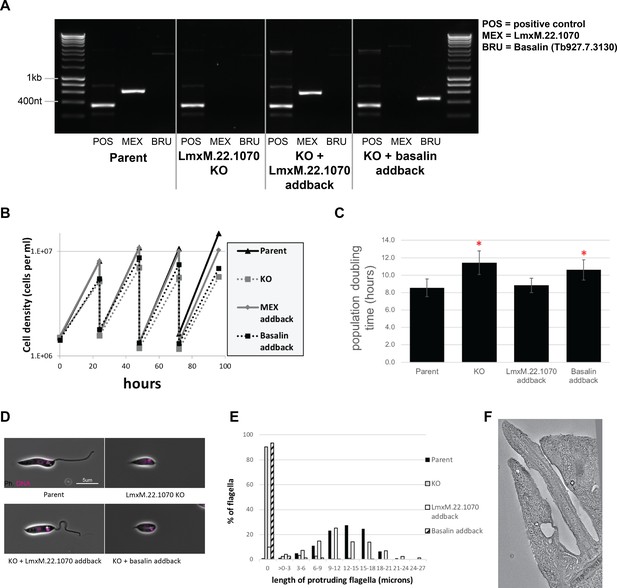
Phenotypic analysis of Leishmania mexicana LmxM.22.1070 knockout cells.
(A) PCR confirms the loss of LmxM.22.1070 in a knockout clone and subsequent addback of LmxM.22.1070 and basalin. (B) LmxM.22.1070 knockout cells grow more slowly than the parental cells. Transfection of a episome addback vector containing LmxM.22.1070 rescues this phenotype, but basalin does not. (C) The doubling times of the populations were calculated based on the data points over 4 days. The asterisk indicates a statistically significant difference compared to the parental cells (T test, p=0.005). (D) Parent and LmxM.22.1070 addback cells have the wildtype morphology, having an elongated cell body and a long flagellum. In contrast, knockout cells and basalin addback cells have a short, round cell body and most cells do not have a flagellum that is visible outside the flagellar pocket of the cell. (E) Cytoskeletons prepared from the four cell lines were stained with the TAT anti tubulin antibody to measure the length of the portion of the flagellum that protrudes beyond the flagellar pocket. ~90% of knockout and basalin addback cells did not have a flagellum that extended beyond the flagellar pocket, and those flagella that did tended to be shorter than the wild type cells. (E) A virtual slice through a Leishmania LmxM.22.1070 KO tomogram shows that no central pair is evident in the flagellum.
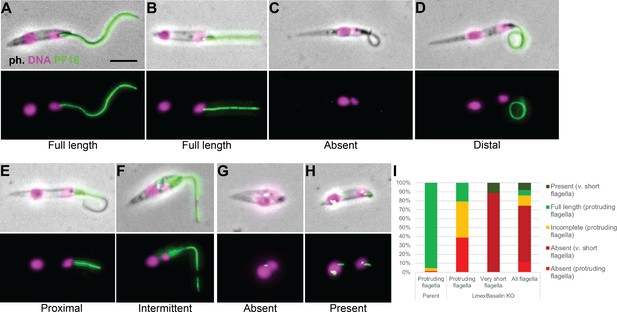
Incorporation of PF16 into LmxM.22.1070 knockout cells.
LmxM.22.1070 was knocked out in Leishmania cells expressing a single allele of PF16 tagged with a C terminal eYFP and cytoskeletons were analysed by fluorescence microscopy. (A) A parental and (B) a KO cell with PF16 incorporated into the entire length of the axoneme. KO cells with (C) PF16 absent from the entire external axoneme, or (D-F) with incomplete PF16 incorporation. A KO cell where PF16 was either (G) absent or (H) present from the region of the cell corresponding to the proximal flagellum. Axoneme incorporation of PF16 was quantified I n (I) log stage cultures (parent N = 152, KO external flagellum = 62, KO no external flagellum = 152, KO total = 214).
-
Figure 10—source data 1
Quantification of PF16 axoneme incorporation in LmxM.22.1070 KO cells.
- https://doi.org/10.7554/eLife.42282.026
Videos
An RNAi screen for paralysed flagella identifies basalin.
https://doi.org/10.7554/eLife.42282.002LmxM.22.1070 knockout Leishmania mexicana cells are mostly paralysed with little or no flagellum visible.
https://doi.org/10.7554/eLife.42282.021Additional files
-
Source code 1
Python script to correct chromatic aberration using multi-chromatic reference beads.
- https://doi.org/10.7554/eLife.42282.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42282.028





