Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease
Figures
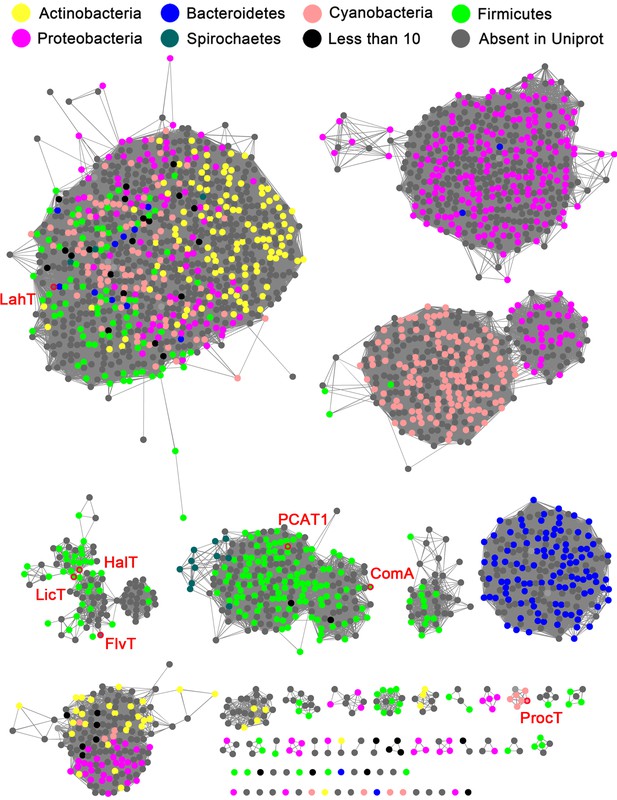
Sequence similarity network (SSN) of select full length AMS/PCAT proteins.
Alignment cutoff of at least 45% sequence identity was applied to separate the clusters. The nodes representing LahT homolog sequences are colored by their corresponding phylum. Nodes of several characterized LahT homologs are marked by red circles and are labeled. The SSN tool draws sequences from UniProt; To increase the coverage of the network, additional sequences not in UniProt were added manually (grey nodes).
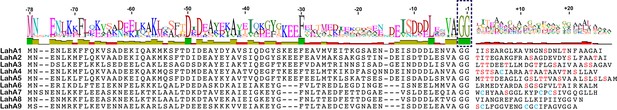
(A) Precursor peptides encoded in Lachnospiraceae C6A11 highlighting the conservation in the leader peptide with a sequence conservation logo (Crooks et al., 2004).
The double Gly motif is boxed.
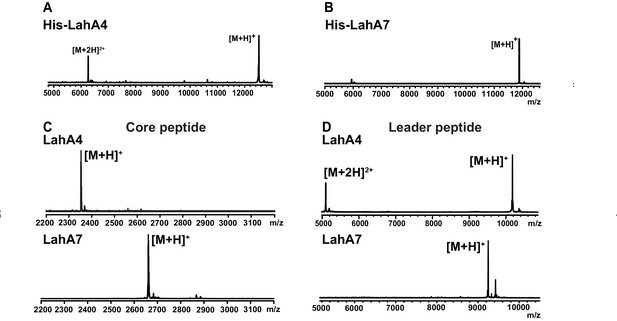
MALDI ToF MS analysis of proteolytic leader peptide removal of two LahA substrates catalyzed by LahT150.
(A–B) MALDI ToF MS analysis of full length N-terminally hexahistidine tagged LahA4 and LahA7. (C) MALDI ToF MS analysis of the core peptides of LahA4 and LahA7 after LahT150 cleavage. Core peptide masses are [M + H]+: LahA4 (calcd 2352.2; obsd 2352.0), LahA7 (calcd 2660.3; obsd 2659.8). (D) MALDI TOF MS analysis of the leader peptides of LahA4 and LahA7 after LahT150 cleavage. Leader peptide average masses are [M + H]+: LahA4-leader (calcd 10188.8, obsd 10187.5), LahA7-leader (calcd 9246.9, obsd 9248.7). For five additional LahA substrates, see Figure 3—figure supplement 1.
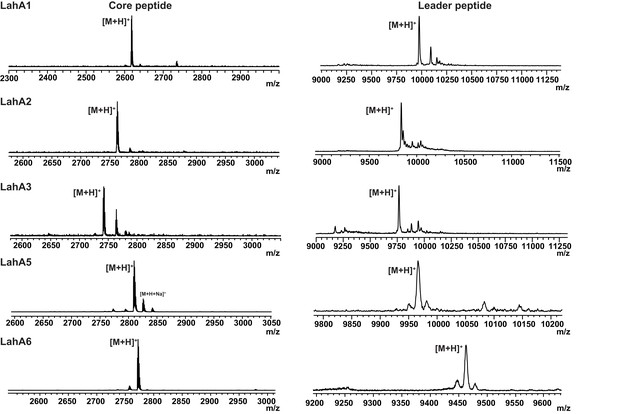
LahT150 cleaves all LahA peptides.
MALDI ToF MS analysis of LahA peptides treated with LahT150. Core peptide ([M + H]+) masses: LahA1 (theor 2616.4; obs 2616.5). LahA2 (theor 2761.4; obs 2762.0). LahA3 (theor 2740.4; obs 2740.7). LahA5 (theor 2811.4; obs 2810.8). LahA6 (theor 2771.4; obs 2771.4). Leader peptide average [M + H]+ masses: LahA1-leader (theor 9973.6, obs 9973.3), LahA2-leader (theor 9827.6, obs 9830.1), LahA3-leader (theor 9756.5, obs 9751.2); LahA5-leader (theor 9964.8, obs 9969.1), LahA6-leader (theor 9458.6, obs 9462.9).
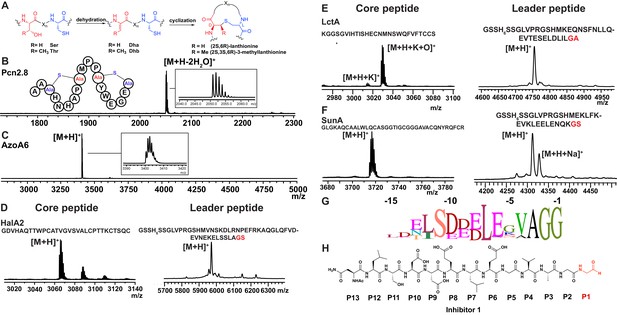
(A) Illustration of the posttranslational modifications in lanthipeptides.
Serine and threonine residues are dehydrated by a lanthionine synthetase, resulting in dehydroalanine (Dha) and dehydrobutyrine (Dhb). The synthetase then catalyzes the Michael type addition of neighboring cysteine residues to the dehydrated residues. (B) Removal of the leader peptide of posttranslationally modified ProcA2.8 monitored by MALDI-TOF MS. Core peptide (two-fold dehydrated) [M + H]+: calcd 2050.8, obsd 2050.9. For four additional ProcA substrates, see Figure 4—figure supplement 1. (C) In vitro leader peptide removal of AzoA6 bearing an N-terminal maltose binding protein tag. Core peptide [M + H]+: calcd 3399.9, obsd 3400.4. For two additional AzoA substrates, see Figure 4—figure supplement 2. (D–F) MALDI TOF MS analysis of LahT150 catalyzed cleavage of the RiPP precursor peptides HalA2, LctA and SunA. Core peptide masses (left panels): HalA2 (calcd 3064.4; obsd 3064.6); LctA (calcd [M + H]+ 3011.3 and [M + H + O]+ 3027.3; obsd 3011.4 and 3027.4); SunA (calcd 3718.7; obsd 3718.6). Leader peptide ([M + H]+) masses (right panels): HalA2-leader peptide (calcd avg. 5969.5; obsd 5969.5); LctA-leader peptide (calcd avg. 4754.2; obsd 4754.6); SunA-leader peptide (calcd avg. 4311.7, obsd 4311.2). (G) Sequence conservation logo (Crooks et al., 2004) showing the frequency of each amino acid (height of the letter) at the C-terminus of the 49 leader peptides in Figure 4—figure supplement 2. (H) Structure of peptide aldehyde inhibitor 1 based on the ProcA2.8 leader peptide.
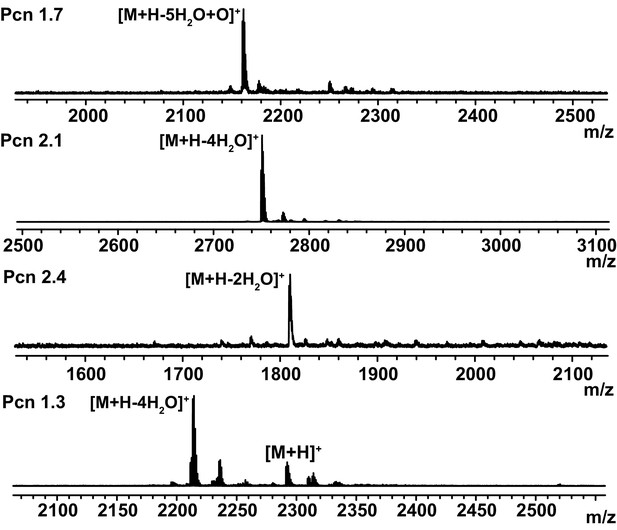
Tests of LahT150 substrate tolerance with posttranslationally modified peptides.
MALDI ToF MS analysis of a selection of ProcM-modified ProcA peptides treated with LahT150. Core peptide products (Pcns) and their ([M + H]+) masses are shown (for sequences see Figure 4—figure supplement 2): Pcn1.7 (theor 2167.1; obs 2166.8). Pcn2.1 (theor 2750.2; obs 2749.9). Pcn2.4 (theor 1808.9; obs 1809.3. Pcn2.8 (cald 2050.8; obs 2051.0). Pcn1.3 (theor 2214.0; obs 2214.5).
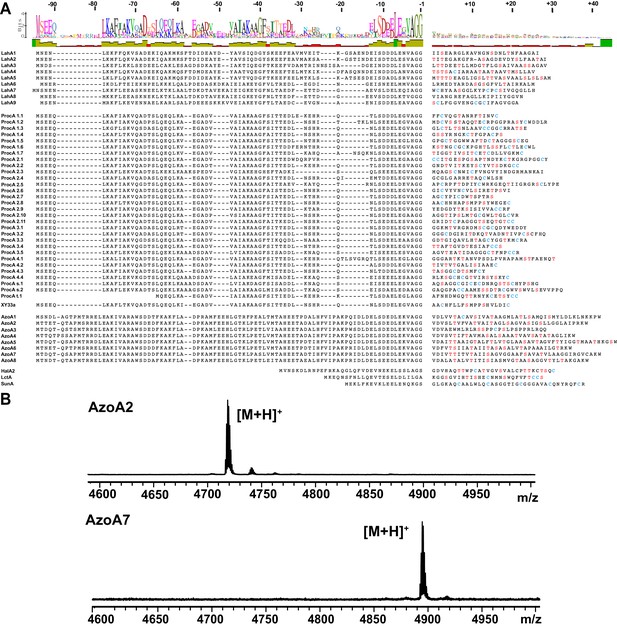
Expanding LahT150 substrate tolerance to non-cognate peptides.
(A) MALDI ToF MS analysis of the LahT150-catalyzed cleavage of MBP-tagged-AzoA2 and MBP-tagged-AzoA7. Both peptides have a C-terminal Asp-Ala-His6 added to the native core peptide sequence to improve their ionization; without these tags, the core peptides ionize poorly. Core peptide [M + H]+ masses: AzoA2 (theor 4718.5, obs 4717.7), AzoA7 (theor 4895.6, obs 4894.8). (B) Sequence alignment for LahA, ProcA, XY33a, and AzoA peptides show strong conservation in the C-terminal 12 amino acids of the leader peptide and very divergent core peptides with no detectable homology. LctA, HalA2 and SunA have low homology to all other peptides but are cleaved by LahT150.
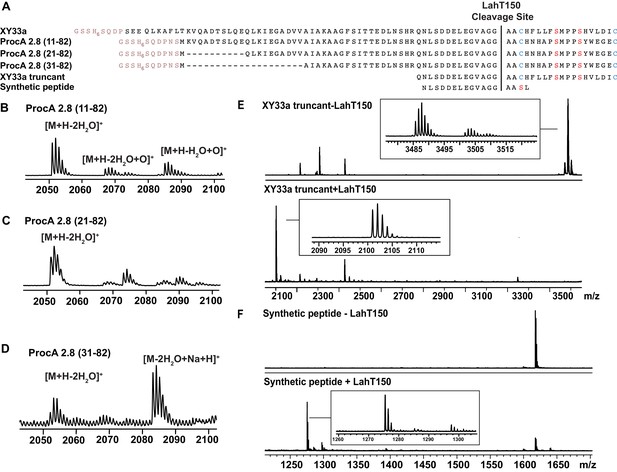
Determination of a minimum sequence for LahT150 catalysis.
(A) Sequence alignment of XY33a, ProcA2.8 (11–82), (21-82 , 31-82) and the XY33a-trypsin generated truncant. (B–E) MALDI ToF MS analysis of the products of N-terminally truncated ProcA2.8 treated with LahT150. LahT150 cleaves all three truncated mutants. ProcA2.8 core peptide mass [M + H-2H2O]+: (theor 2050.8; obs 2051.1). (F) XY33a was treated with trypsin to generate the XY33a truncant shown in panel (B), then the trypsin was inactivated by boiling before treatment with LahT150. LahT150 processed the trypsin-generated XY33a truncant. Core peptide masses: ([M + H]+): XY33a-trypsin truncant (theor 3485.6; obs 3485.7); XY33a-core peptide (theor 2101.0; obs 2100.9). (G) MALDI ToF MS analysis of the synthetic peptide in Figure 4—figure supplement 3A without LahT150 (top panel) and with LahT150 (bottom panel). Synthetic peptide [M + H]+ (theor 1617.8, obs 1617.8), Cleaved synthetic peptide N-terminal fragment [M + H]+ (theor 1275.6, obs 1275.5 Da).
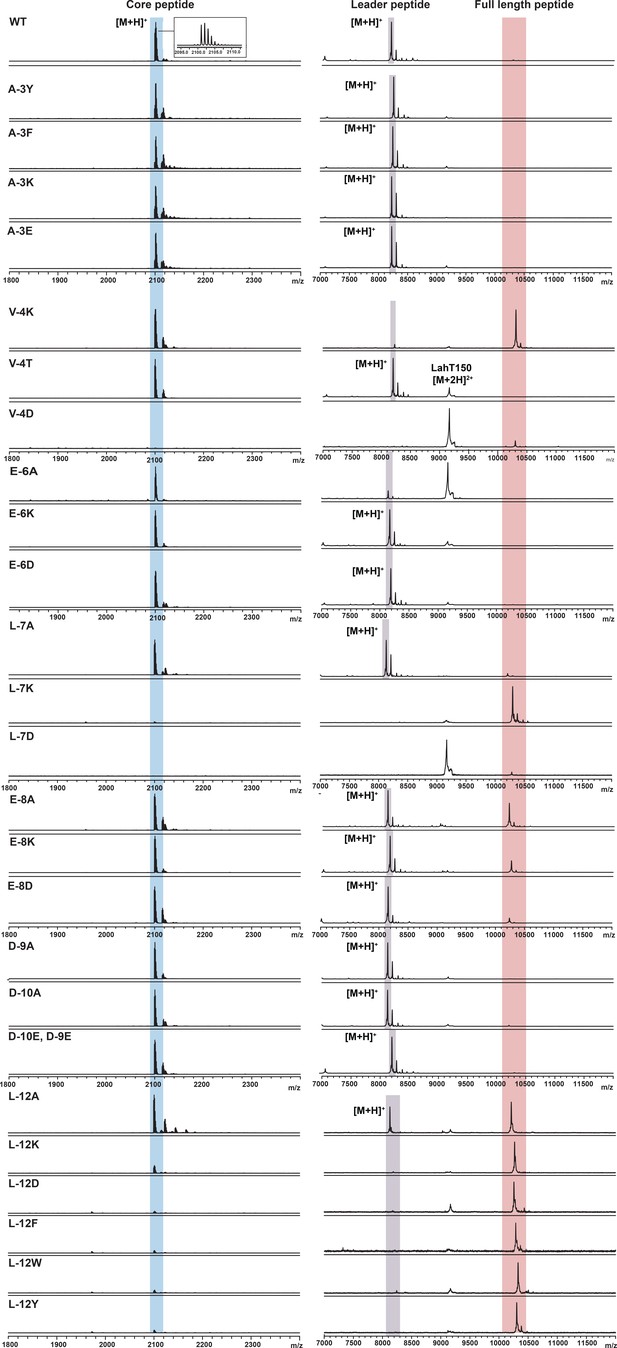
Assessment of the importance of individual amino acids in the leader peptide for LahT150 catalysis.
LahT150 catalyzed cleavage reactions of XY33a wild-type and leader peptide mutants. XY33a-core peptide mass [M + H]+ theor 2100.0; obs 2100.9–2101.1 in all spectra.
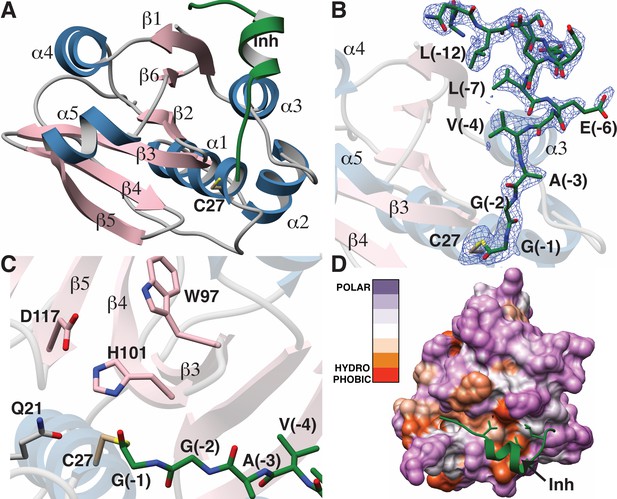
Structure of the LahT147-peptide aldehyde complex.
(A) Overall structure of the complex showing the orientation of the peptide aldehyde (colored in green and labeled as Inh). (B) Simulated annealing difference Fourier map (calculated without the coordinates for Cys27 and the peptide aldehyde and shown at 2.3 σ) superimposed on the coordinates of the complex. (C) Close-up view of the active site showing residues implicated in catalysis. (D) Hydropathy analysis of LahT147 (based on the Kyte and Doolittle scale [Kyte and Doolittle, 1982]) superimposed in a color scheme onto a surface rendering of the final structure. Note that Val−4, Leu−7, and Leu−12 of the leader are positioned in suitable hydrophobic pockets. The figure was generated using the Chimera software package (Pettersen et al., 2004).
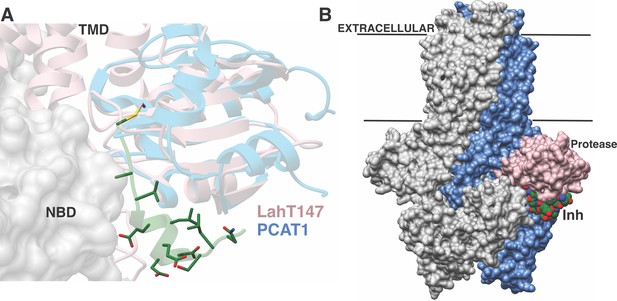
Structure-based superposition of LahT147 and PCAT1.
(A) Close-up view of the LahT147-inhibitor complex structure superimposed on the crystal structure of full-length PCAT1. Note that the leader sequence directs the core peptide ‘cargo’ into the transmembrane domain (TMD) and is flanked by the nucleotide-binding domain (NBD). (B) Overall structure of the PCAT1 dimer with one monomer colored grey and the other monomer blue and pink showing the relative orientations of the protease domain and the inhibitor. Binding of the peptide cargo is poised to stabilize the interdomain interactions in the full-length transporter.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (XY33a_V-4K_gene) | XY33a_V-4K_gene | IDT. Representative of other purchased XY33a mutant genes (see Table 5) | XY33a_V-4K_gene | 5 ng/μL stock solution (1 μL) used as amplification template |
| Strain, strain background (Escherichia coli BL21 (DE3)-T1R) | E. coli BL21 (DE3)-T1R | Sigma Aldrich B2935 | BL21 (DE3)-T1R | |
| Strain, strain background (Lachnospiraceae C6A11) | Lachnospiraceae C6A11 | Dr. William Kelly (AgResearch, New Zealand) | Lachnospiraceae C6A11 | |
| Strain, strain background (Escherichia coli Rosetta 2 (DE3)) | E. coli Rosetta 2 (DE3) | Novagen Catalog no. 71400–3 | E. coli Rosetta 2 (DE3) | |
| Strain, strain background (Azospirillum sp. B510 (JCM 14679)) | Azospirillum sp. B510 (JCM 14679) | JCM Riken http://www.jcm.riken.jp/cgi-bin/jcm/jcm_number?JCM=14679 | Azospirillum sp. B510 (JCM 14679) | |
| Transformed construct (pETDuet LahT150) | pETDuet-LahT150 | this work | pETDuet LahT150 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet XY33a) | pRSFDuet-XY33a | PMID: 22574919 | pRSFDuet XY33a | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet XY33a A-3Y) | pRSFDuet XY33a A-3Y | this work. Representative XY33a mutant | pRSFDuet XY33a A-3Y | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA1) | pRSFDuet-LahA1 | this work | pRSFDuet LahA1 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA2) | pRSFDuet-LahA2 | this work | pRSFDuet LahA2 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA3) | pRSFDuet-LahA3 | this work | pRSFDuet LahA3 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA4) | pRSFDuet-LahA4 | this work | pRSFDuet LahA4 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA5) | pRSFDuet-LahA5 | this work | pRSFDuet LahA5 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA6) | pRSFDuet-LahA6 | this work | pRSFDuet LahA6 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet LahA7) | pRSFDuet-LahA7 | this work | pRSFDuet LahA7 | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pET28-MBP-AzoA2) | pET28-MBP-AzoA2 | this work | pET28-MBP-AzoA2 | 150 ng/μL stock solution (1 μL) used in E. coli Rosetta 2 (DE3) transformation |
| Transformed construct (pET28-MBP-AzoA3) | pET28-MBP-AzoA3 | this work | pET28-MBP-AzoA3 | 150 ng/μL stock solution (1 μL) used in E. coli Rosetta 2 (DE3) transformation |
| Transformed construct (pET28-MBP-AzoA6) | pET28-MBP-AzoA6 | this work | pET28-MBP-AzoA6 | 150 ng/μL stock solution (1 μL) used in E. coli Rosetta 2 (DE3) transformation |
| Transformed construct (pET28-MBP-AzoA7) | pET28-MBP-AzoA7 | this work | pET28-MBP-AzoA7 | 150 ng/μL stock solution (1 μL) used in E. coli Rosetta 2 (DE3) transformation |
| Transformed construct (pRSFDuet ProcA 2.8 (MCSI) - ProcM (MCSII)) | pRSFDuet ProcA2.8 (MCSI) - ProcM (MCSII) | PMID: 22574919 | pRSFDuet ProcA 2.8 (MCSI) - ProcM (MCSII) | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet ProcA 1.7 (MCSI) - ProcM (MCSII)) | pRSFDuet ProcA1.7 (MCSI) - ProcM (MCSII | PMID: 22574919 | pRSFDuet ProcA 1.7 (MCSI) - ProcM (MCSII | 50 ng/μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet ProcA 2.1 (MCSI) - ProcM (MCSII)) | pRSFDuet ProcA2.1 (MCSI) - ProcM (MCSII | this work | pRSFDuet ProcA 2.1 (MCSI) - ProcM (MCSII | 50 ng/ μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet ProcA 2.4 (MCSI) - ProcM (MCSII)) | pRSFDuet ProcA2.4 (MCSI) - ProcM (MCSII) | this work | pRSFDuet ProcA 2.4 (MCSI) - ProcM (MCSII) | 50 ng/ μL stock solution (1 μL) used in E. coli BL21 transformation |
| Transformed construct (pRSFDuet ProcA 1.3 (MCSI) - ProcM (MCSII)) | pRSFDuet ProcA1.3 (MCSI) - ProcM (MCSII) | this work | pRSFDuet ProcA 1.3 (MCSI) - ProcM (MCSII) | 50 ng/ μL stock solution (1 μL) used in E. coli BL21 transformation |
| Sequence- based reagent | Benzonase Endonuclease | EMD Millipore Catalog no. 1.01656.001 | Benzonase | |
| Sequence- based reagent | EcoRI-HF | New England Biolabs R3101S | EcoRI | |
| Sequence- based reagent | BamHI-HF | New England Biolabs R3136S | BamHI | |
| Sequence- based reagent | NotI-HF | New England Biolabs R3189S | Not1 | |
| Sequence- based reagent | HindIII-HF | New England Biolabs R3104S | HindIII | |
| Recombinant protein | XY33a | PMID: 29507389 | XY33a | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | XY33a A-3Y | this work; representative XY33a mutant | XY33a A-3Y | recombinant substrate peptide mutant tested with LahT150 |
| Recombinant protein | LahA1 | this work | LahA1 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA2 | this work | LahA2 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA3 | this work | LahA3 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA4 | this work | LahA4 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA5 | this work | LahA5 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA6 | this work | LahA6 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahA7 | this work | LahA7 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | AzoA2 | this work | MBP-AzoA2 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | AzoA3 | this work | MBP-AzoA3 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | AzoA6 | this work | MBP-AzoA6 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | AzoA7 | this work | MBP-AzoA7 | recombinant substrate peptide tested with LahT150 |
| Recombinant protein | LahT150 | this work | LahT150 | protease domain of LahT |
| Recombinant protein | Pcn 2.8 | PMID: 22574919 | Pcn 2.8 | recombinant posttranslationally modified ProcA2.8 substrate peptide tested with LahT150 |
| Recombinant protein | Pcn 1.7 | PMID: 22574919 | Pcn 1.7 | recombinant posttranslationally modified ProcA1.7 substrate peptide tested with LahT150 |
| Recombinant protein | Pcn 2.1 | this work | Pcn 2.1 | recombinant posttranslationally modified ProcA2.1 substrate peptide tested with LahT150 |
| Recombinant protein | Pcn 2.4 | this work | Pcn 2.4 | recombinant posttranslationally modified ProcA2.4 substrate peptide tested with LahT150 |
| Recombinant protein | Pcn 1.3 | this work | Pcn 1.3 | recombinant posttranslationally modified ProcA1.3 substrate peptide tested with LahT150 |
| Minimum peptide substrate | Synthetic peptide | Genscript | Synthetic peptide | synthetic minimal substrate peptide tested with LahT150 |
| Commercial kit | QIAprep Spin Miniprep kit | Qiagen catalog no. 27106 | QIAprep Spin Miniprep kit | |
| Commercial kit | QIAquick Gel Extraction kit | Qiagen catalog no. 28115 | QIAquick Gel Extraction kit | |
| Commercial kit | Gibson Assembly | New England Biolabs E2611S | Gibson Assembly | |
| Chemical compound | TCEP (Tris (2-Carboxyethyl) phosphine hydrochloride) | Goldbio Catalog ID TCEP | TCEP | |
| Chemical compound | Terrific Broth granulated | Fisher Scientific BP97285 | TB | |
| Chemical compound | Glycerol | Fisher Scientific BP-229–4 | Glycerol | |
| Chemical compound | Dextrose | Fisher Scientific BP350500 | Glucose or dextrose | |
| Chemical compound | kanamycin monosulfate, USP grade | Goldbio Catalog ID K-120 | kanamycin | |
| Software, algorithm | Adobe Illustrator CS6 | Adobe | Adobe Illustrator | |
| Software, algorithm | FlexAnalysis 3.4 (Bruker Daltonik GmbH) | Bruker Daltonik GmbH | FlexAnalysis 3.4 (Bruker Daltonik GmbH) | Mass spectrometry data processing |
| Other | Clontech His60 Ni Superflow resin | Clontech Catalog no. 636660 | Clontech His60 Ni Superflow resin | Used for gravity purification of all recombinant proteins except LahT150 |
| Other | GE Healthcare HisTrap HP | GE Healthcare 17524701 | 5 mL HiTrap Ni Chelating column | Used for FPLC purification of recombinant LahT150 |
Calculated and observed MALDI ToF [M + H]+ masses for the leader peptides in Figure 4—figure supplement 4. n.d., not detected.
https://doi.org/10.7554/eLife.42305.013| [M + H]+ | WT | V-4K | V-4T | V-4D | E-6A | E-6K | E-6D |
|---|---|---|---|---|---|---|---|
| Calcd | 8169.8 | 8227.0 | 8199.8 | 8229.8 | 8139.8 | 8168.9 | 8183.8 |
| Obsd | 8169.3 | 8229.9 | 8201.4 | n.d. | 8137.8 | 8170.0 | 8183.1 |
| L-7A | L-7K | L-7D | E-8A | E-8K | E-8D | ||
| Calcd | 8127.7 | 8212.9 | 8199.8 | 8111.8 | 8168.9 | 8155.8 | |
| Obsd | 8126.2 | n.d. | n.d. | 8111.0 | 8167.7 | 8157.5 | |
| D-9A | D-10A | D-9E,D-10E | A-3Y | A-3F | A-3K | A-3E | |
| Calcd | 8125.8 | 8125.8 | 8197.9 | 8261.9 | 8245.9 | 8226.9 | 8227.9 |
| Obsd | 8125.4 | 8127.0 | 8196.7 | 8261.1 | 8246.5 | 8225.8 | 8226.8 |
| L-12A | L-12K | L-12D | L-12F | L-12W | L-12Y | ||
| Calcd | 8127.7 | 8184.8 | 8171.7 | 8203.8 | 8242.8 | 8184.8 | |
| Obsd | 8126.0 | n.d. | n.d. | n.d. | 8241.0 | n.d |
Data collection, phasing and refinement statistics.
https://doi.org/10.7554/eLife.42305.014| LahT-inhibitor 1 complex | PCMBS | ||
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | |
| Unit cell (a,b,c,β) | 37.9, 119.4, 76.5, 93.8 | 37.3, 119.8, 83.5, 112.8 | |
| Resolution | 76.4–1.98 (1.985–1.98) | 59.9–2.04 (2.05–2.04) | |
| Total reflections | 239,058 | 124,854 | |
| Unique reflections | 47,187 | 21,494 | |
| Rsym (%)* | 0.102 (0.727) | 0.090 (0.690) | |
| I/σ(I)* | 9.3 (2.1) | 12.9 (2.5) | |
| Completeness (%)* | 99.8 (99.8) | 99.9 (100) | |
| Redundancy | 5.1 (5.1) | 5.9 (6.0) | |
| Refinement | |||
| Resolution (Å) | 50.0–2.0 | ||
| No. reflections | 43,389 | ||
| Rwork / Rfree† | 23.4/26.8 | ||
| Number of atoms | |||
| Protein | 4479 | ||
| Inh | 352 | ||
| Water | 123 | ||
| B-factors | |||
| Protein | 37.6 | ||
| Inh | 34.5 | ||
| Water | 35.9 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.015 | ||
| Bond angles (°) | 1.81 | ||
-
*Highest resolution shell is shown in parenthesis.
†R-factor = Σ(|Fobs|-k|Fcalc|)/Σ |Fobs|and R-free is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
Primers used in the generation of LahA constructs.
Homology with vector backbone is displayed as lowercase letters.
| Primer Name | Sequence 5’−3’ |
|---|---|
| LahA1_fp | accatcatcaccacagccaggatccgaattcgaACGAGAATTTAGAGAAGTTTTTTCAGA |
| LahA1_rp | ttctgttcgacttaagcattatgcggccgcAGATTGCTCCTGCAGCGAAATTGGTAAG |
| LahA2_fp | accatcatcaccacagccaggatccgaattcgaACGAGAATTTAAAGATGTTTTTGCAGA |
| LahA2_rp | ttctgttcgacttaagcattatgcggccgcTTAGATTGCTGTTGCAGCGAAAAGGGAAT |
| LahA3_fp | accatcatcaccacagccaggatccgaattcgaATGATAGTTTAAAAGAGTTTTTGAA |
| LahA3_rp | ttctgttcgacttaagcattatgcggccgcTTAGACGGCTCCGGCTGACGATGCCGCAA |
| LahA4_fp | accatcatcaccacagccaggatccgaattcgaACGAGAATTTAAAGATGTTTTTACAGA |
| LahA4_rp | ttctgttcgacttaagcattatgcggccgcTTAAACCGCAAGTAAACTCATCGTTACAGC |
| LahA5_fp | accatcatcaccacagccaggatccgaattcgaACGAGAATCTCAAGCTATTTTTACAA |
| LahA5_rp | ttctgttcgacttaagcattatgcggccgcTTACATTGCCGATAATGATAATGATAATGC |
| LahA6_fp | accatcatcaccacagccaggatccgaattcgaATGAAAGGATAAAAGATTTATTTACCG |
| LahA6_rp | ttctgttcgacttaagcattatgcggccgcTTACATAAGTGCCTTTCTTATTGCAGTAAG |
| LahA7_rp | accatcatcaccacagccaggatccgaattcgaACGAGAACTTGAAGAAATTCCTGGAGG |
| LahA7_fp | ttctgttcgacttaagcattatgcggccgcTTATGAAGCAATCCTTGACCAACTATTGA |
Primers used in the cloning of AzoA constructs.
Homology with vector backbone is displayed as lowercase letters.
| Primer name | Sequence 5'−3' |
|---|---|
| AzoA2 fwd | aaaGGATCCatgacaaccgaaacgcaaacc |
| AzoA2 rev | aaaGCGGCCGCctaccattttctgggaatggccaag |
| AzoA3 fwd | caatggacggtGGATCCGatgacagaccaaacccagtccacatcc |
| AzoA3 rev | cggaaacagccAAGCttactgttgtcgcaaacgcggtggtga |
| AzoA6 fwd | aaaggacttcgGGATCCgatgacaaatgaaacgcagcccacc |
| AzoA6 rev | ttatgggatcCAAGCTTctaccatttcctcgttccgagaatggc |
| AzoA7 fwd | caatggacccaGGATCCgatgacagaccaaacgcagtccgcc |
| AzoA7 rev | catggacatcCAAGCTTctaccattttgcacacacccccctgat |
| MBP-AzoA G1 | aataaggagatataccatgGGCAGCAGCCATCATCATCATC |
| MBP-AzoA G2 | TGGCTGCTGCCcatggtatatctccttattaaagttaaacaaaattatttctacagggg |
| MBP-AzoA2 G3 | CTGTACTTCCAATCCatgacaaccgaaacgcaaaccgcc |
| MBP-AzoA2 G4 | cgtttcggttgtcatGGATTGGAAGTACAGGTTCTCAGATCCACGC |
| MBP-AzoA3 G3 | CTGTACTTCCAATCCatgacagaccaaacccagtccac |
| MBP-AzoA3 G4 | ggtttggtctgtcatGGATTGGAAGTACAGGTTCTCAGATCCACGC |
| MBP-AzoA6 G3 | CTGTACTTCCAATCCatgacaaatgaaacgcagccc |
| MBP-AzoA6 G4 | cgtttcatttgtcatGGATTGGAAGTACAGGTTCTCAGATCCACGC |
| MBP-AzoA7 G3 | CTGTACTTCCAATCCatgacagaccaaacgcagtccgcc |
| MBP-AzoA7 G4 | gcgtttggtctgtcatGGATTGGAAGTACAGGTTCTCAGATCCACGC |
| AzoA2CHis G1 | tctaGTGATGGTGATGGTGATGTGCATCccattttctgggaatggccaagc |
| AzoA2CHis G2 | GATGCACATCACCATCACCATCACtagaagcttgcggccgcataatgcttaagtcg |
| AzoA3CHis G1 | tctaGTGATGGTGATGGTGATGTGCATCctgttgtcgcaaacgcggtggtg |
| AzoA3CHis G2 | GATGCACATCACCATCACCATCACtagaagcttgcggccgcataatgcttaagtcg |
| AzoA7CHis G1 | tctaGTGATGGTGATGGTGATGTGCATCccattttgcacacacccccctgattccacc |
| AzoA7CHis G2 | GATGCACATCACCATCACCATCACtagaagcttgcggccgcataatgcttaagtcg |
Primers and synthetic genes used in the cloning of XY33a constructs.
Mutations are shown in bold font. Homology with vector backbone is displayed as lowercase letters.
| Primer or synthetic gene name | Sequence (5’−3’) |
|---|---|
| XY33A_fp | catcaccatcatcaccacagccaggatccGTCTGAAGAGCAACTGAAGGC |
| XY33A_rp | gtacaatacgattactttctgttcgacttaagcattatTTAGCAAATATCGAGGACGTG |
| RSF_fp | Gcaggcgtttttccatagg |
| RSF_rp | Ctggcttgagcgtcgatttttg |
| XY33a_D-9A_fp | CGCCAAAATCTGTCTGAA GCA AGCTGGAAGGTGTGGC |
| XY33a_D-9A_rp | GCCACACCTTCCAGCT TGC TTCAGACAGATTTTGGCG |
| XY33a_D-10A_fp | CGCCAAAATCTGTCT GCA GAAAGCTGGAAGGTGTGGC |
| XY33a_D-10A_rp | GCCACACCTTCCAGCTTTC TGC AGACAGATTTTGGCG |
| XY33a_D-10E,D-9E_fp | CGCCAAAATCTGTCT GAA GAA AGCTGGAAGGTGTGGCTG |
| XY33a_D-10E,D-9E_rp | CAGCCACACCTTCCAGCT TTC TTC AGACAGATTTTGGCG |
| XY33a_E-8A_fp | CAAAATCTGTCTGATGAT GCA CTGGAAGGTGTGGCTGGG |
| XY33a_E-8A_rp | CCCAGCCACACCTTCCAG TGC ATCATCAGACAGATTTTG |
| XY33a_E-8K_fp | CAAAATCTGTCTGATGAT AAA CTGGAAGGTGTGGCTGGG |
| XY33a_E-8K_rp | CCCAGCCACACCTTCCAG TTT ATCATCAGACAGATTTTG |
| XY33a_E-8D_fp | CAAAATCTGTCTGATGAT GAT CTGGAAGGTGTGGCTGGG |
| XY33a_E-8D_rp | CCCAGCCACACCTTCCAG ATC ATCATCAGACAGATTTTG |
| XY33a_L-7A_fp | CTGTCTGATGATGAG GCA GAAGGTGTGGCTGGGG |
| XY33a_L-7A_rp | CCCCAGCCACACCTTC TGC CTCATCATCAGACAG |
| XY33a_L-7K_fp | CTGTCTGATGATGAG AAA GAAGGTGTGGCTGGGG |
| XY33a_L-7K_rp | CCCCAGCCACACCTTC TTT CTCATCATCAGACAG |
| XY33a_L-7D_fp | CTGTCTGATGATGAG GAT GAAGGTGTGGCTGGGG |
| XY33a_L-7D_rp | CCCCAGCCACACCTTC ATC CTCATCATCAGACAG |
| XY33a_E-6A_fp | GTCTGATGATGAGCTG GCA GGTGTGGCTGGGGGAG |
| XY33a_E-6A_rp | CTCCCCCAGCCACACC TGC CAGCTCATCATCAGAC |
| XY33a_E-6K_fp | GTCTGATGATGAGCTG AAA GGTGTGGCTGGGGGAG |
| XY33a_E-6K_rp | CTCCCCCAGCCACACC TTT CAGCTCATCATCAGAC |
| XY33a_E-6D_fp | GTCTGATGATGAGCTG GAT GGTGTGGCTGGGGGAG |
| XY33a_E-6D_rp | CTCCCCCAGCCACACC ATC CAGCTCATCATCAGAC |
| XY33a_V-4K_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGTAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATCTGTCTGATGATGAGCTGGAAGGTAAAGCTGGGGGAGCGG CCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_V-4T_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGTAGGCTTCTCGATTACCACAGAAGACCTAAACTC TCATCGCCAAAATCTGTCTGATGATGAGCTGGAAGGTACCGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_V-4D_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAA CAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGTAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATCTGTCTGATGATGAGCTGGAAGGTGATGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_A-3Y_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCAT CGCCAAAATCTGTCTGATGATGAGCTGGAAGGTGTGTATGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACG TCCTCGATATTTGCTAA |
| XY33a_A-3F_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATCTGTCTGATGATGAGCTGGAAGGTGTGTTTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_A-3E_gene | TCTGAAGAGCAACTGAAGGCATTCCTCA CCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATG TTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTC TCATCGCCAAAATCTGTCTGATGATGAGC TGGAAGGTGTGGAAGGGGGAGCGGCCT GTCATTTCCTTCTTTTCTCTATGCCTCC ATCCCACGTCCTCGATATTTGCTAA |
| XY33a_A-3K_gene | TCTGAAGAGCAACTGAAGGCATTCCTCA CCAAAGTTCAAGCCGATACTTCACTACAG GAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCT CATCGCCAAAATCTGTCTGATGATGAGCTGGAAGGTGTGAAAGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCC TCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12A_gene | TCTGAAGAGCAACTGAAGGCATTCCTC ACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGC TGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGAC CTAAACTCTCATCGCCAAAATGCGTC TGATGATGAGCTGGAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12K_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATAAATCTGATGATGAGCTGGAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCC TTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12D_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATGATTCTGATGATGAGCTGGAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12F_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATTTTTCTGATGATGAGCTGGAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12W_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATGTTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATTGGTCTGATGATGAGCTGGAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
| XY33a_L-12Y_gene | TCTGAAGAGCAACTGAAGGCATTCCTCACCAAAGTTCAAGCCGATACTTCACTACAGGAACAGTTAAAGATAGAAGGAGCTGATG TTGTAGCCATTGCCAAAGCTGCAGGCTTCTCGATTACCACAGAAGACCTAAACTCTCATCGCCAAAATTATTCTGATGATGAGCTG GAAGGTGTGGCTGGGGGAGCGGCCTGTCATTTCCTTCTTTTCTCTATGCCTCCATCCCACGTCCTCGATATTTGCTAA |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42305.018





