Interferon lambda 4 impacts the genetic diversity of hepatitis C virus
Figures
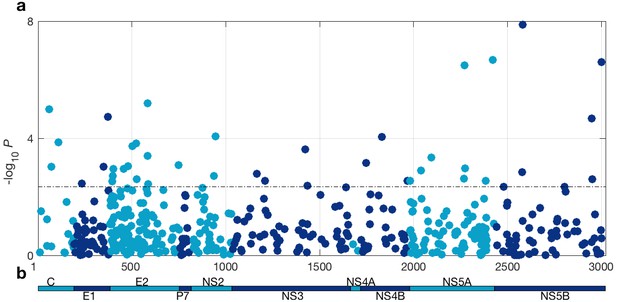
HCV genome-wide association study with IFNL4 SNP rs12979860 genotypes (CC vs. non-CC).
(a) Manhattan plot. The dashed line indicates 5% FDR. At this level 42 sites on the virus polyprotein are significantly associated with IFNL4 SNP. (b) Schematic of the HCV polyprotein.
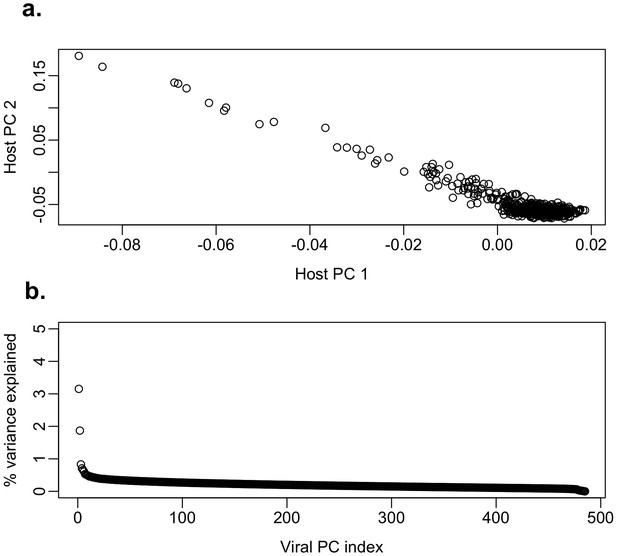
Host and viral principal components.
(a) Host first and second PCs. (b) Proportion of variance explained by the viral principal components (PCs). The first and second PCs explain 3% and 2% of variation in the nucleotide sequences respectively which indicates there is little clustering of the viral sequences. This is also consistent with the virus phylogenetic tree, which is star-like.
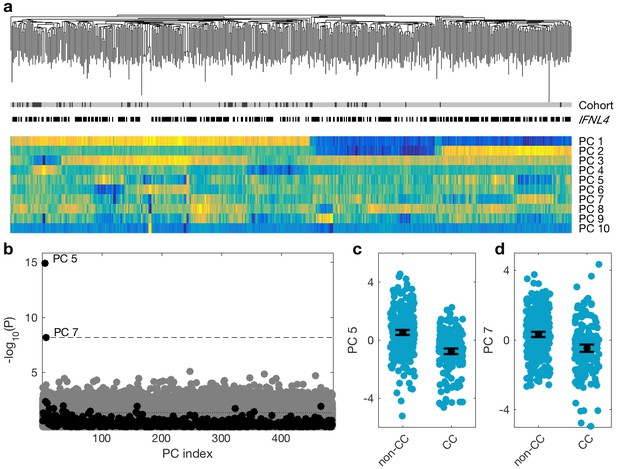
Association between viral PCs and IFNL4 SNP rs12979860 genotypes in the combined cohort (N = 485).
(a) Virus phylogenetic tree, cohort (black EAP, grey BOSON), IFNL4 SNP rs12979860 (CC white, non-CC black) and the first 10 viral PCs (the colours are mapped such that dark blue represents the smallest number and bright yellow represents the largest number for each PC). (b) P-value of univariate association tests between viral PCs and the host SNPs. Black and grey dots are for association tests between the viral PCs and the IFNL4 SNP rs12979860 and 500 SNPs with minor allele frequency (MAF) similar to IFNL4 SNP rs12979860 MAF, respectively. Dashed line shows the 10% FDR line and the dotted line shows the nominal significance of p=0.05. Distribution of the fifth (c) and seventh (d) PCs stratified by the host IFNL4 SNP rs12979860 genotypes. Black dot and lines show the mean and 95% confidence interval for each group.
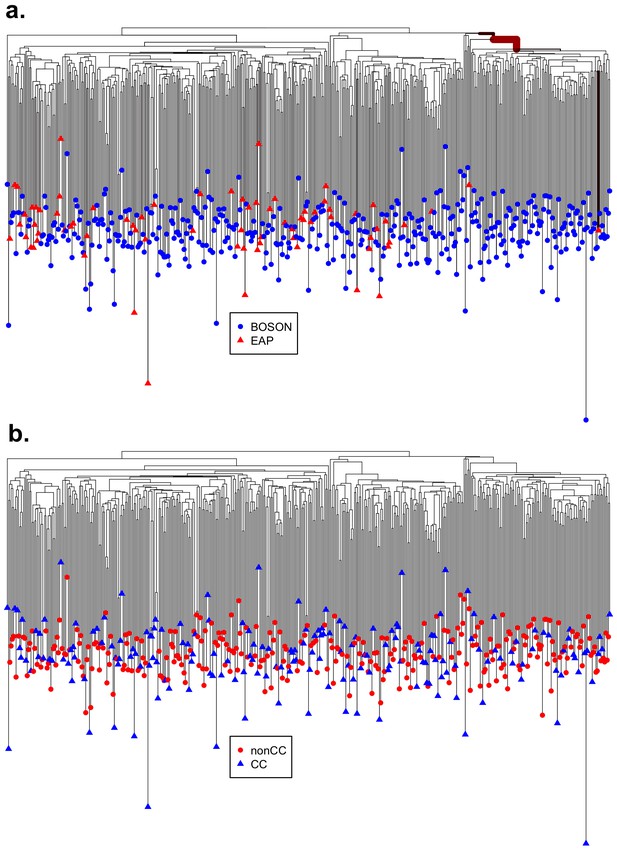
Distribution of (a) cohort from which the sequences were obtained (BOSON N = 411 or EAP N = 74) and (b) host IFNL4 SNP rs12979860 genotypes (CC or non-CC) on the virus phylogenetic tree.
The thickness and redness of the branches are proportional to the posterior probability that the distribution of the trait of interest on the tips of the tree is different under that clade. (a) Bayes factor of the alternative (where there is one or more branches which have a different distribution of sequences from the BOSON and EAP cohorts) to null model (where the distribution of sequences from the BOSON and EAP cohorts is the same everywhere on the tree) is 249, indicating that the alternative model is supported. There is a branch (thick and red) on the tree, representing a clade within which there are very few EAP sequences as well as sequences from UK patients in the BOSON cohort. (b) There is no evidence that any part of the tree has a distinct distribution in terms of the host IFNL4 SNP rs12979860 genotypes, the estimated Bayes factor for alternative model (where there is one or more branches which have a different distribution of host IFNL4 genotypes) to null model (where the distribution of IFNL4 SNP rs12979860 genotypes is the same everywhere on the tree) is 1.1 which indicates that the null model that host IFNL4 SNP rs12979860 genotypes are randomly distributed on the virus tree is better supported.
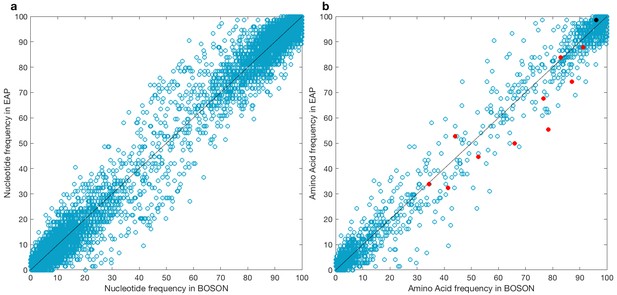
Viral allele frequencies in the BOSON (N = 411) and EAP (N = 74) cohorts.
(a) Viral nucleotide and (b) amino acid frequencies in the BOSON and EAP cohorts. The red dots represent the 10 amino acids previously reported to be associated with IFNL4 genotype. The black dot represents position 2576, which was identified in our previous study as a site associated with IFNL4 genotype (Ansari et al., 2017) but was not tested in the present study.
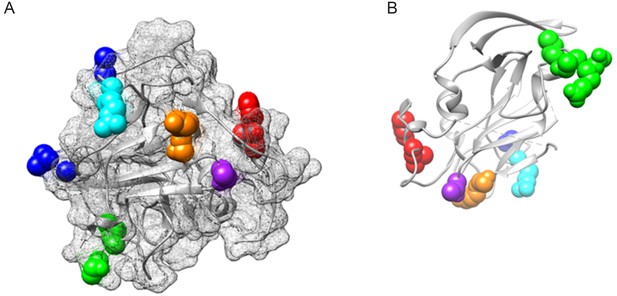
IFNL4-associated residues on the core E2 structure.
The core E2 structure (PDB 4MWF) is shown in grey, (A) shows the protein surface as a mesh (B) shows the ribbon structure. IFNL4 associated residues are highlighted as follows: L438 and F442 in Epitope 2 (red), K500 and S501 in orange, R521 in cyan, A524 and L546 in Epitope 3 (blue), T558 in purple and D576, N577, T578 and L580 in the igVR (green).
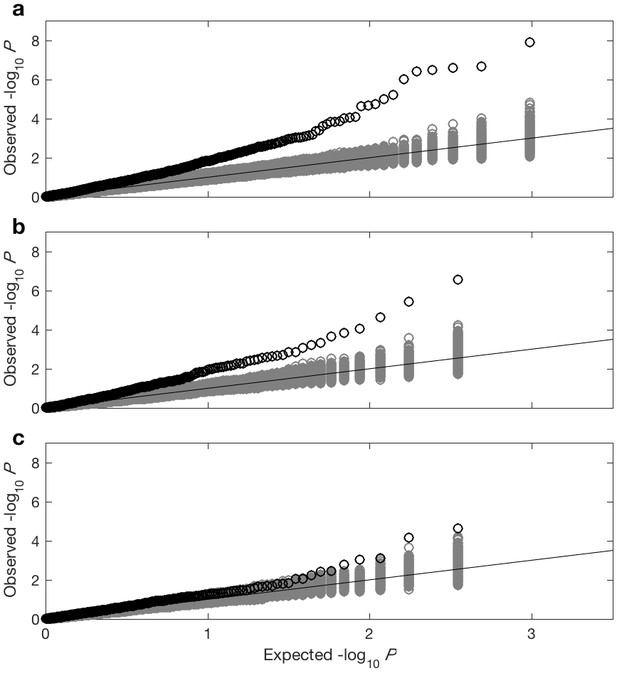
QQ-plots for association studies between viral amino acids and viral codons and host IFNL4 SNP rs12979860 and 500 host SNPs chosen across the human genome with a minor allele frequency (MAF) similar to the IFNL4 SNP rs12979860 MAF.
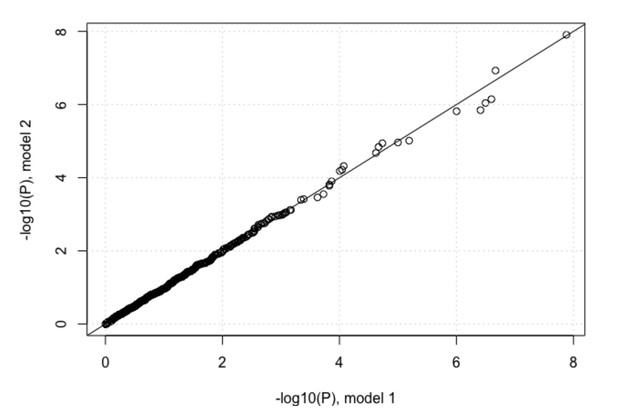
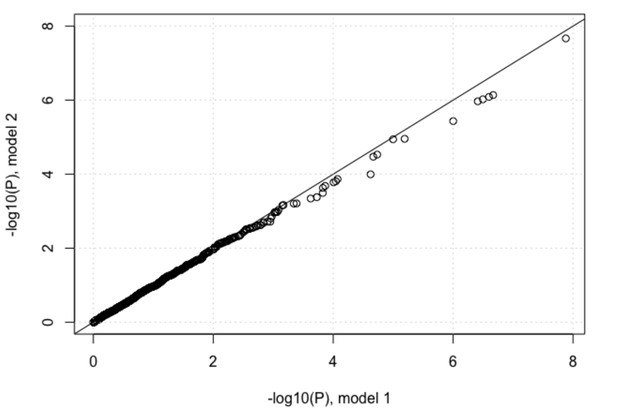
QQ-plots for association tests between the host SNPs and viral amino acid (a) host SNPs and change from the most common viral codon to (b) non-synonymous codons and (c) synonymous codons. First two viral PCs and first three host PCs were used as covariates in all three analyses. The black circles show the QQ-plot for the virus GWAS against IFNL4 SNP rs12979860 and the grey circles show the QQ-plot for the virus GWASs against 500 frequency-matched SNPs.
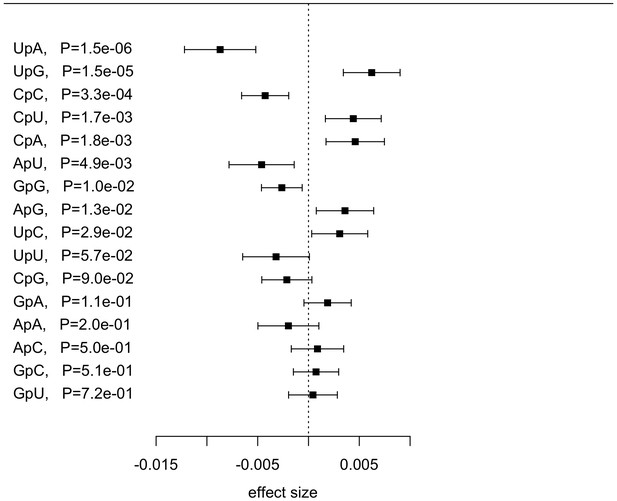
Effect size (beta) of IFNL4 SNP rs12979860 genotypes (CC vs. non-CC) on the proportion of dinucleotide frequencies in the combined cohort (N = 485).
The effect size is measured for non-CC relative to CC group.
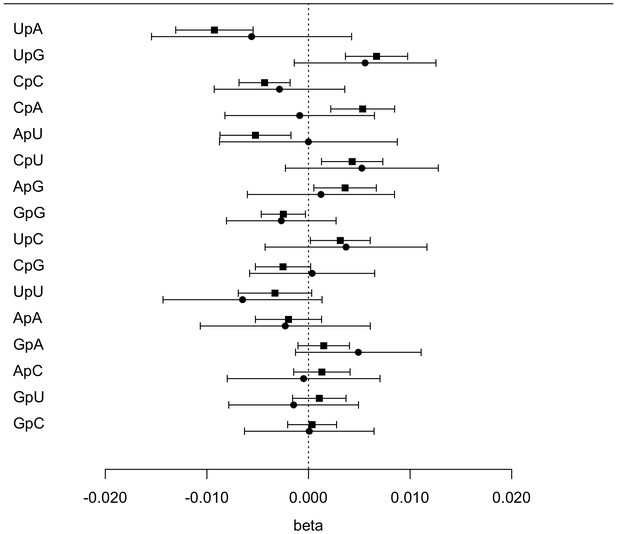
Effect size (beta) of IFNL4 SNP rs12979860 genotypes (CC vs. non-CC) on the proportion of dinucleotide frequencies in the BOSON (N = 411) and EAP (N = 74) cohorts.
The effect size is measured for non-CC relative to CC group. The square indicates the effect size in the BOSON cohort and the circle indicates the effect size in the EAP cohort.
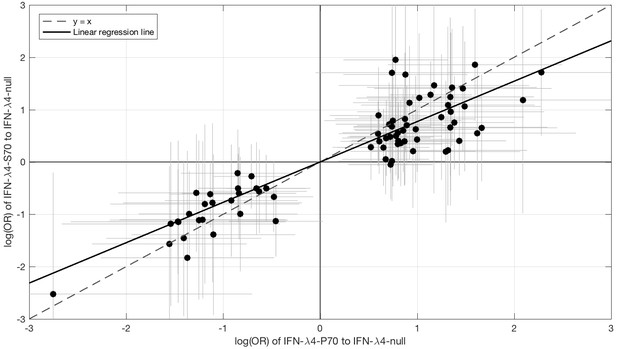
Comparison of the effect sizes (log(OR)) of host IFN-λ4 variants (IFN-λ4-S70 and IFN-λ4-P70 relative to IFN-λ4-Null) on HCV-encoded amino acids.
The circles show the log(OR) estimates and the grey lines indicate the 95% confidence intervals. The dashed line is the y = x line which has a slope of one. The solid black line shows the linear regression line, which has a slope of 0.77 that is significantly different from one (y = x line, p=9.6×10−7).
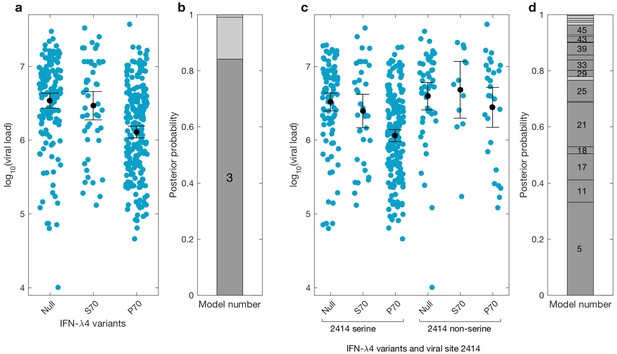
Bayesian model comparison of effect sizes of IFNL-λ4 variants on viral load in the BOSON cohort (N = 411).
(a) Pretreatment viral load stratified by the host IFN-λ4 variants. The black dots and lines indicate the mean and 95% confidence interval (CI) for each group. (b) The posterior probability of the five tested models from (a) stacked on top of each other (from model 1 to model 5; posterior probabilities of models 1, 2 and 5 are too small to be labelled on this plot). Models where the posterior probability is higher or lower than the prior probability are coloured as dark grey and light grey respectively. Only model 3 (as indicated) has a posterior probability bigger than its prior probability and it assumes that the mean viral load is identical in IFN-λ4-Null and IFN-λ4-S70 groups and different from the mean viral load of IFN-λ4-P70 group. (c) Viral load stratified by the host IFNL-λ4 variants and the presence and absence of serine at the viral amino acid site 2414. The black dots and lines indicate the mean and 95% CI for each group. (d) The posterior probability of the 58 tested models from (c) stacked on top of each other (from model 1 to model 58; only models where the posterior probability is higher than the prior probability are labelled on this plot). Models where the posterior probability is higher or lower than the prior probability are coloured as dark grey and light grey respectively. Model 5 has the highest posterior probability and it assumes that the mean viral load is only different in ‘IFN-λ4-P70 + 2414 serine’ group and identical in other groups.
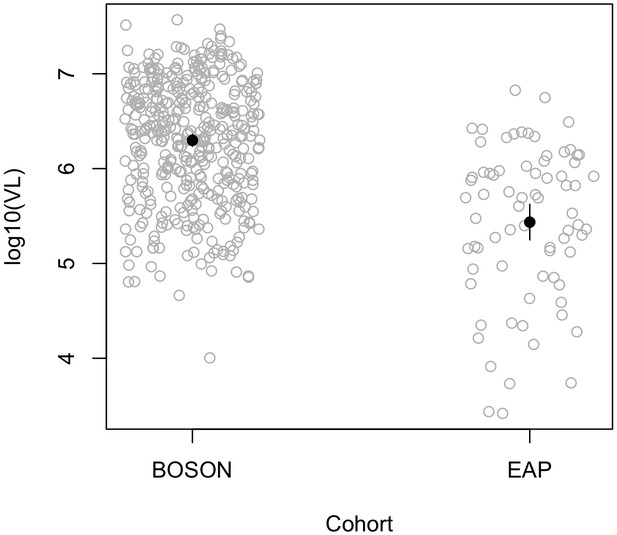
Pre-treatment viral load stratified by cohort.
The viral load in the EAP cohort (N = 74) is significantly lower than that of the BOSON cohort (N = 411) (p=1.49×10−13).
Additional files
-
Supplementary file 1
Demographic, clinical and genetic characteristics of the BOSON and EAP cohorts.
- https://doi.org/10.7554/eLife.42463.014
-
Supplementary file 2
Host IFNL4 SNP rs12979860 association with HCV amino acids at a 10% FDR.
We used logistic regression to test for association between host IFNL4 SNP (CC vs. non-CC) and presence and absence of amino acids. We included the first two viral and the first three host PCs as covariates. Only amino acids that were present in at least 20 samples were tested (977 amino acids in 471 sites). For each associated site, we have reported all amino acid with a count of >= 20 in reducing frequency order and highlighted the most associated amino acid in bold. The amino acid frequency in CC and nonCC patients, the P-value, log(OR), the standard error and q-value are reported for the most associated amino acid at the site.
- https://doi.org/10.7554/eLife.42463.015
-
Supplementary file 3
P-value of Fisher’s exact test for enrichment or depletion of the association signals in HCV proteins and HLA-restricted epitopes.
- https://doi.org/10.7554/eLife.42463.016
-
Supplementary file 4
Associations between HCV amino acids and the 500 frequency-matched host SNPs at a 5% FDR.
Note that the FDR was calculated independently for each viral GWAS against each host SNP. All the significant associations for each viral GWAS (against each of the 500 frequency matched host SNP) are shown in this table.
- https://doi.org/10.7554/eLife.42463.017
-
Supplementary file 5
Host IFNL4 SNP rs12979860 association with changes from the most common codon to non-synonymous codons in HCV at a 10% FDR.
We used logistic regression to test for association between host IFNL4 SNP (CC vs. non-CC) and codon changes. We included the first two viral and the first three host PCs as covariates. Only codons at which there were at least 20 synonymous and 20 non-synonymous codons for the most common codon at the site (348 codon sites across the HCV coding sequence) were included in the analysis.
- https://doi.org/10.7554/eLife.42463.018
-
Supplementary file 6
Host IFNL4 SNP rs12979860 association with changes from the most common codon to synonymous codons in HCV at a 10% FDR.
We used logistic regression to test for association between host IFNL4 SNP (CC vs. non-CC) and codon changes. We included the first two viral and the first three host PCs as covariate. Only codons at which there were at least 20 synonymous and 20 non-synonymous codons for the most common codon at the site (348 codon sites across the HCV coding sequence) were included in the analysis.
- https://doi.org/10.7554/eLife.42463.019
-
Supplementary file 7
IFNL4 haplotype combination and predicted protein for host SNPs rs117648444 and rs368234815 in the EAP (N = 74) and BOSON (N = 411) cohorts.
- https://doi.org/10.7554/eLife.42463.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42463.021