High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms
Figures
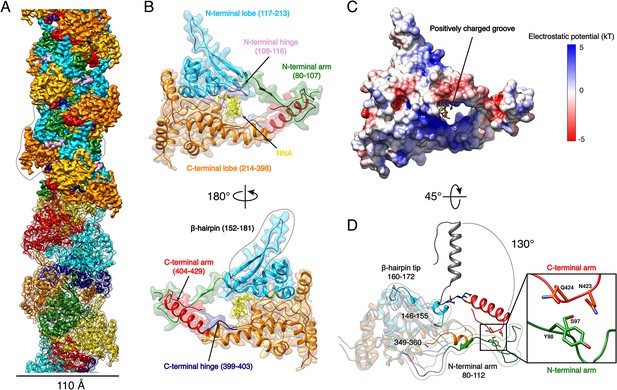
HTNV-NC structure.
(A) HTNV-NC helical organisation. The upper part of the cryo-EM map is coloured per NP domain, whereas the lower part is shown as transparent and coloured per NP protomer. The NP surrounded by dotted lines corresponds to Figure 1B orientation. (B) Domain organisation of one monomer extracted from the NC helical assembly. Each domain is coloured as in Figure 1A upper part. The newly described β-hairpin (residues 152–181) is labelled and surrounded by dotted lines. (C) Electrostatic surface representation of HTNV-NP monomer. (D) Superimposition of monomeric NPcore (Olal and Daumke, 2016) (in gray) and NP from the present structure (coloured as in Figure 1A,B). The Ctarm rotation is highlighted and newly built elements are labelled, shown as non-transparent and surrounded by dotted lines. A close up view of the Ctarm/Ntarm interaction is shown.
-
Figure 1—source data 1
Cryo-EM data collection, refinement and validation statistics.
- https://doi.org/10.7554/eLife.43075.007
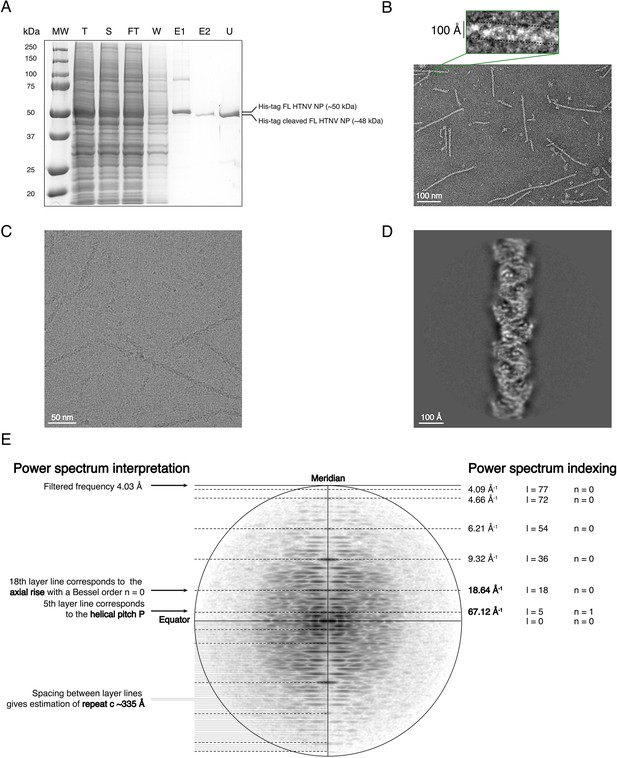
Purification, EM analysis and symmetry determination of HTNV-NC.
(A) SDS-PAGE gel of HTNV-NC purification. T: total fraction, S: soluble fraction, FT: flow through of the 1st Ni-NTA purification, W: wash of the 1st Ni-NTA purification, E1: elution of the 1st Ni-NTA purification, E2: supernatant after TEV cleavage and 2nd Ni-NTA purification, U: concentrated protein after ultra-centrifugation. (B,C) Negative stain (B) and cryo electron micrographs (C) of HTNV-NC. Scale bar is indicated. (D) 2D class average. (E) 2D class average power spectrum, indexing and interpretation enabling symmetry determination.
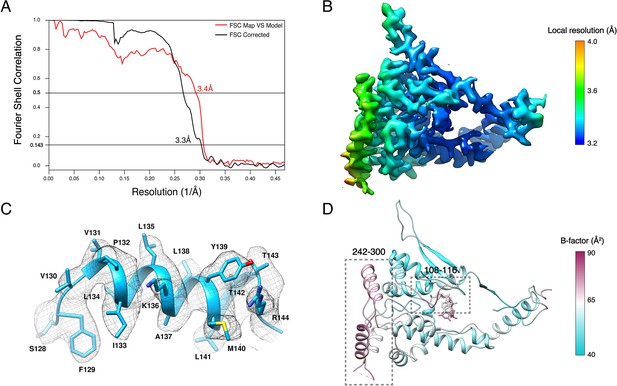
Resolution of HTNV-NC structure.
(A) The gold-standard Fourier shell correlation (FSC) of masked map indicates a resolution of 3.3 Å with the FSC = 0.143 criteria. The FSC between the map and the model is 3.4 Å according to FSC = 0.5 criterion. (B) Local resolution. (C) Zoom-up view of a representative α-helix that shows clear side chains densities illustrating the quality of the map. (D) The B-factor analysis strongly correlates with the local resolution analysis (B) and shows that the N-terminal hinge and the solvent-exposed region containing residues 242–300 are flexible.
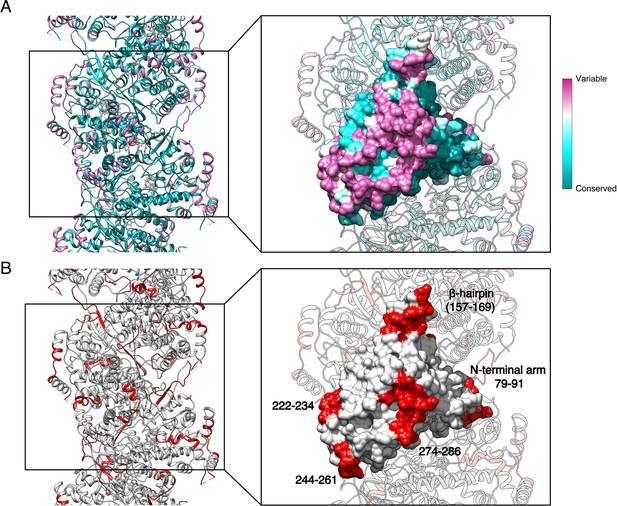
Specific antigenic sites are localised in variable regions of the NC surface.
(A) Surface conservation mapping of HTNV-NC. Regions localised towards NC interior, in particular regions involved in RNA-binding and NP-NP oligomerisation are conserved, whereas HTNV-NC exposed regions are more variable. (B) Epitopes involved in serotype-specific and cross-reactive recognition of HTNV-NC determined in (Tischler et al., 2008) are shown in red. They correspond to surface-exposed regions of HTNV-NC corroborating their necessary accessibility.
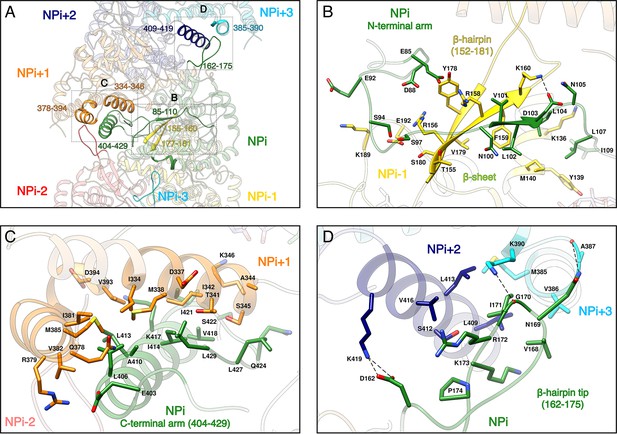
NP-NP interactions.
(A) General view. The filament orientation corresponds to RNA direction from 5’ to 3’. Each NP protomer is coloured differently. Interacting regions of NPi with subunits ranging from. NPi-3 to NPi+3 are shown as non-transparent. Positions of Figure 2B,C,D close-up views are indicated with dotted lines. (B) NPi Ntarm binding site in NPi-1. Residues 101–103 from NPi Ntarm form a β-sheet with β-strands 155–160 and 177–181 from NPi-1. Hydrogen bonds are shown as black dotted lines. (C) NPi Ctarm binding site in NPi+1. (D) Binding of NPi β-hairpin tip (residues 162–175) on NPi+2 (residues 409–419) and NPi+3 (residues 385–390). NPi I171 is plugged into a hydrophobic pocket formed by L409 and L413 of the NPi+2 C-terminal helix and M385 and K390 sidechains of the NPi+3 C-terminal lobe. Hydrogen bonds between the residue pairs N169-A387 and G170-K390 from NPi and NPi+3 respectively, further stabilise this interaction contributing to the rigidification of HTNV-NC.
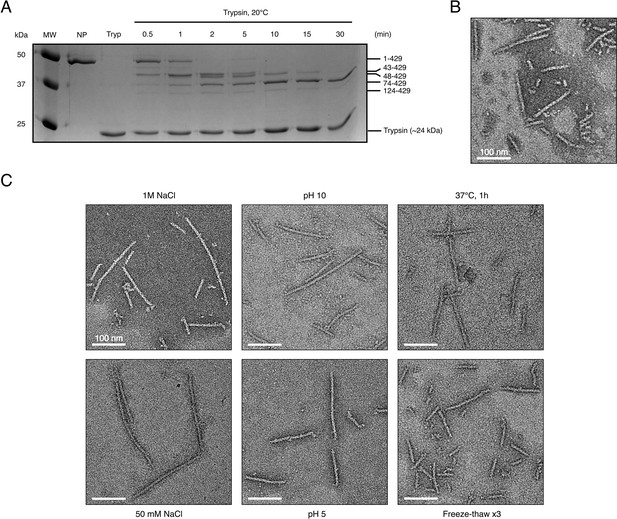
Limited proteolysis and stability of recombinant HTNV-NCs.
(A) SDS-Page analysis of trypsin digested HTNV-NC. MW represents the molecular weight markers. NC were incubated with trypsin (Tryp) at 20°C for different duration from 0 (NP) to 30 min. After 30 min, a stable construct containing residues 74 to 429 is obtained. Presence of intermediate limited proteolysis fragments containing residues 43–429 and 48–429 strongly suggests that the N-terminal residues 1–73 are not disordered and might correspond to a coiled-coil as observed by Boudko et al. (2007). (B) Negative stain EM micrograph of truncated construct NP74-429 displays NC similar to the ones of full-length NP. (C) Negative stain micrographs of full-length HTNV-NCs incubated in different pHs and salt concentration as indicated. NC fibers stay stable in harsh conditions. Consecutive freeze-thaw breaks NC in small rigid parts. Scale bars are indicated.
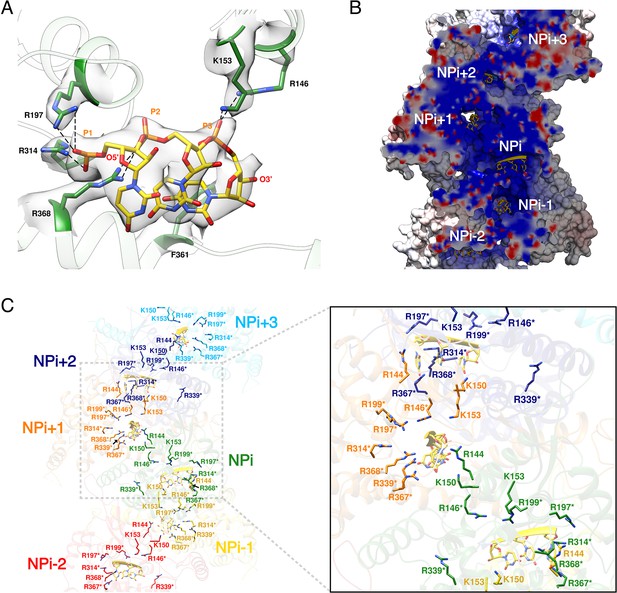
HTNV-NC RNA binding site.
(A) RNA binding mode of 3 nucleotides. EM density of RNA and RNA-binding residues are displayed in transparent grey. Hydrogen bonds are shown as dotted lines. (B) Cut-away view of NC electrostatic potential showing the continuous positively-charged groove. RNA nucleotides are displayed. (C) RNA-binding residues are shown as sticks and coloured per subunit. RNA-binding residues defined in Guo et al. (2016), namely R146*, R197*, R199*, R314*, R339*, R367*, R368* are labelled with stars, while RNA-binding residues identified in the present structure, namely R144, K150, K153 are shown without stars.
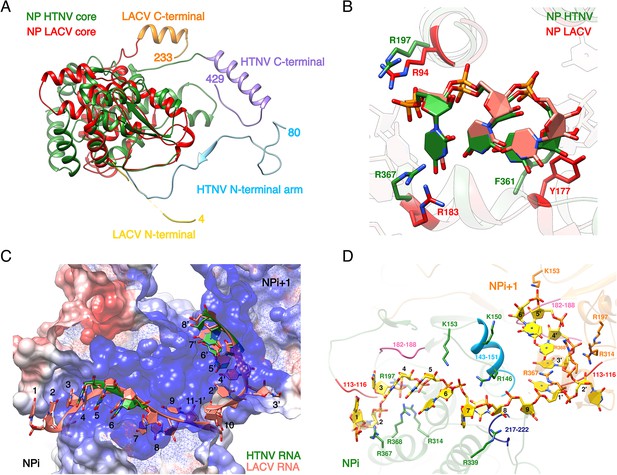
Comparison of LACV and HTNV-NP/NC, RNA-binding model.
(A) Superimposition of HTNV and LACV NPcores. HTNV and LACV Ntarms and Ctarms contain the same secondary structures but LACV arms are significantly shorter and differ in orientation. (B) HTNV-NP-bound nucleotides adopt a similar conformation as LACV-NP-bound nucleotides 5 to 7. The nucleotide binding pocket is conserved in HTNV-NP and LACV-NP. (C) Cut-away view of HTNV-NC electrostatic potential. Visible HTNV nucleotides are shown in green. LACV nucleotides position based on superimposition of LACV and HTNV NPcores are shown in salmon. LACV RNA fits reasonably well into HTNV-NC RNA path, however LACV nucleotide 11 from NPi clashes with LACV nucleotide one from NPi+1 showing that each HTNV-NP binds less than 11 nucleotides. LACV nucleotides numbering is indicated. (D) Model of HTNV-NC bound to nine nucleotides. Nucleotides numbering from 1 to 9 is indicated. Residues interacting with riboses and phosphates are shown as sticks. Regions interacting with bases are coloured and specifically labelled.
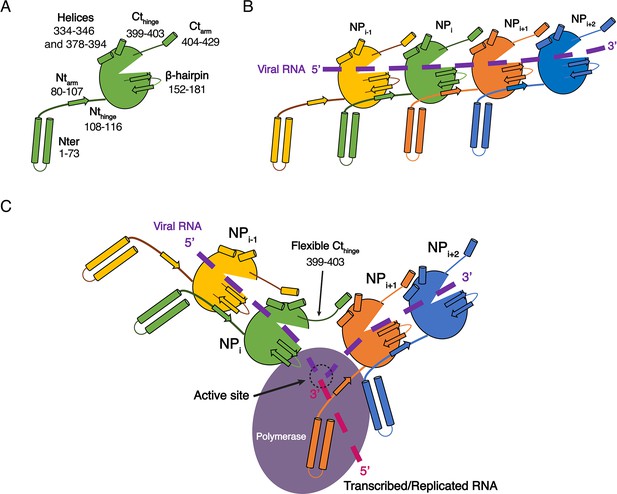
Model of HTNV replication and transcription.
(A) Schematics representation of HTNV-NP. Major secondary structures involved in interprotomer interactions are represented as arrows for β-strand and cylinders for α-helices. RNA binding cavity is represented as a clipped part from the NPcore region (green oval). (B) Schematic representation of RNA binding and NP-NP interactions. RNA is shown as a dotted purple line. Main NP-NP interactions between adjacent subunits are indicated. For clarity, NPi interactions with NPi-2, NPi-3, NPi+2 and NPi+3 are absent from the schematic representation. (C) Replication working hypothesis model which is inspired from Gerlach et al. (2015). The polymerase is shown in purple and the newly transcribed/replicated RNA is indicated as a pink dotted line. The model proposes that the polymerase binds to the flexible N-terminal1-73 region during replication/transcription in order to move along the NC. This would be reminiscent to P and L binding to flexible sNSV C-terminal region. Binding of the polymerase to the N-terminal1-73 region could destabilise the adjacent Ntarm binding. This localised disruption of NP-NP interaction would create a local opening of the NC enabling transient access of the polymerase to few RNA nucleotides for replication/transcription. Such an opening could be possible without disturbing the whole NC as NP to NP contacts are driven not only by the Ntarm but also by the Ctarm. The Ctarm interaction is likely to remain intact even with a local opening of the NC as the C-terminal hinge allows the Ctarm to undergo large rotation.

Purification and negative stain electron microscopy analysis of HTNV NP and L co-expression/co-purification.
(A) SDS-PAGE gel of HTNV NP-L co-expression/co-purification that was done as follow: insect cells were co-infected with two baculoviruses respectively producing NP and L. Purification then consisted in Ni-NTA affinity and S200 gel filtration. (B) Fraction 2 of the SDS-Page gel contains HTNV-NC that are indistinguishable from HTNV-NC purified in the absence of L. (C) Fraction 6 of the SDS-Page gel contains homogeneous HTNV-L and very small HTNV-NC but no interaction is visible.
Bent recombinant NCs seen in some EM micrographs.
Bending localisations are shown with arrows.
Videos
Overview of HTNV-NC structural organisation.
The helical arrangement of HTNV-NC is coloured as in Figure 1A upper part. HTNV-NP domain organisation is coloured as in Figure 1B. Ctarm/Ntarm interaction is highlighted.
Interaction of subunit NPi with six neighbouring subunits.
Subunit NPi (in green) interacts: (i) with subunit NPi-1 (in yellow) via its Ntarm, (ii) with subunit NPi+1 (in orange) via its Ctarm, with subunit NPi+2 (in dark blue) and subunit NPi+3 (in cyan) via its β-hairpin tip. Due to helical symmetry, NPi-3 β-hairpin tip (in light cyan) also interacts with NPi (same interaction as NPi-NPi+3). Consistently, NPi-2 β-hairpin tip (in red) also interacts with NPi (same interaction as NPi-NPi+2).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | |
|---|---|---|---|---|---|
| recombinant DNA reagent | HTNV-NP | Geneart | Synthetic gene | ||
| Peptide, recombinant protein | HTNV-NP | This article | UniProtKB-P05133 | HTNV-NP was obtained by expression in insect cells of the synthetic geneHTNV-NP mentioned above | |
| Software, algorithm | Motioncor2 | doi: 10.1038/nmeth.2472 | http://msg.ucsf.edu/em/software/motioncor2.html | ||
| Software, algorithm | Gctf | doi: 10.1016/j.jsb.2015. 11.003 | RRID:SCR_016500 | https://www.mrc-lmb.cam.ac.uk/kzhang/ | |
| Software, algorithm | Relion2.1 and Relion3 | doi: 10.1016/j.jsb.2012. 09.006 and doi: 10.1101/421123 | RRID:SCR_016274 | https://www2.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page | |
| Software, algorithm | Coot | doi: 10.1107/S090744 4910007493 | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | RCrane | doi: 10.1107/S090744 4912018549 | https://pylelab.org/software/rcrane-readme | ||
| Software, algorithm | PHENIX | doi: 10.1107/S090744 4909052925 | RRID:SCR_014224 | https://www.phenix-online.org/ | |
| Software, algorithm | Chimera | doi: 10.1002/jcc.20084 | RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera/ | |
| Software, algorithm | Haddock | doi: 10.1038/nprot.2010.32 and 10.1016/j.jmb.2015.09.01 | https://haddock.science.uu.nl/ | ||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43075.015





