zGrad is a nanobody-based degron system that inactivates proteins in zebrafish
Figures
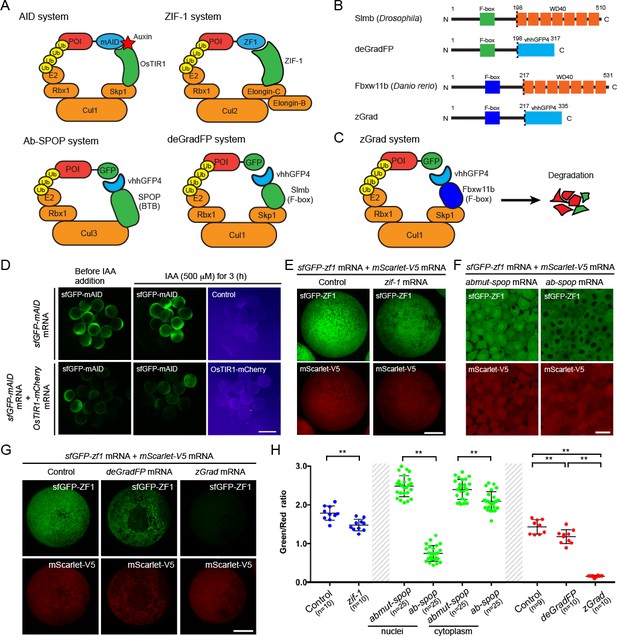
zGrad degrades GFP-tagged proteins in zebrafish.
(A) Overview of degron-based protein degradation systems. POI: protein of interest. (B) Comparison of deGradFP and zGrad fusion proteins. (C) Schematic of zGrad-mediated target protein degradation. (D) Representative images of embryos injected with sfGFP-mAID mRNA (210 pg) only (top) or with sfGFP-mAID mRNA (210 pg) and OsTIR1-mCherry mRNA (42 pg) (bottom) before (left, 8 hpf) and after IAA (500 µM) induction for 3 hr (middle and right, 11 hpf). Red channel is shown as a fire map (right). Scale bar: 1 mm. (E) Representative images of embryos injected with sfGFP-ZF1 mRNA and mScarlet-V5 mRNA only (left) or with sfGFP-ZF1 mRNA and mScarlet-V5 mRNA and zif-1 mRNA (right) at 9 hpf. Scale bar: 200 µm. Note that mScarlet-V5 fluorescence served as an internal control. (F) Single-plane confocal images of cells in embryos injected with sfGFP-ZF1 mRNA, mScarlet-V5 mRNA and abmut-spop mRNA (left) or ab-spop mRNA (right) at 9 hpf. Scale bar: 20 µm. (G) Representative images of embryos injected with sfGFP-ZF1 mRNA and mScarlet-V5 mRNA (left) or with sfGFP-ZF1 mRNA and mScarlet-V5 mRNA and deGradFP mRNA (middle) or zGrad mRNA (right) at 9 hpf. Scale bar: 200 µm. (H) Quantification of control and Zif-1-mediated sfGFP-ZF1 degradation shown in E (blue), Abmut-SPOP control and Ab-SPOP-mediated sfGFP-ZF1 degradation shown in F (green), and deGradFP-mediated and zGrad-mediated sfGFP-ZF1 degradation shown in G (red). Mean, SD and n are indicated. **p<0.01. .
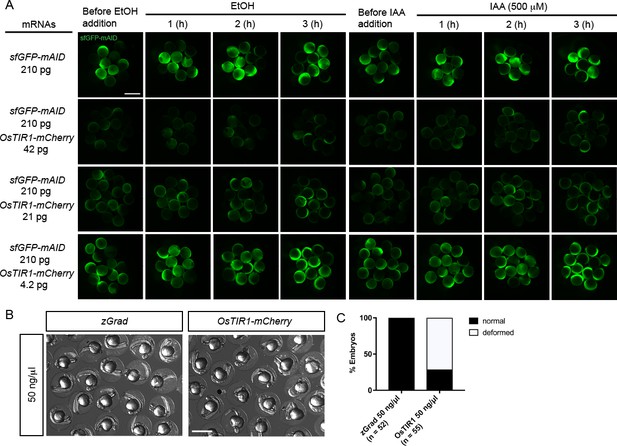
Characterization of the AID system in zebrafish.
(A) Images of embryos injected with sfGFP-mAID mRNA and different amounts of OsTIR1-mCherry mRNA and incubated with 0.2%EtOH (left) or the auxin IAA 500 µM (right) over a 3 hr time course. Scale bar: 1 mm. (B) Images of embryos injected with zGrad mRNA and OsTIR1-mCherry mRNA at 24 hpf. Scale bar: 1 mm. (C) Percentage of deformed embryos among embryos injected with zGrad mRNA and OsTIR1-mCherry mRNA at 24 hpf.
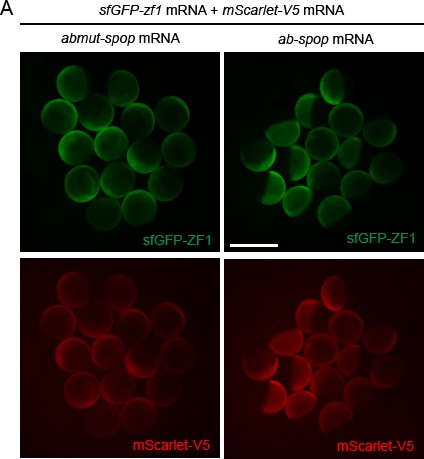
Characterization of the Ab-SPOP system in zebrafish.
(A) Images of embryos injected with sfGFP-ZF1 mRNA, mScarlet-V5 mRNA and abmut-spop mRNA (left) or ab-spop mRNA (right) at 8 hpf. Scale bar: 1 mm.
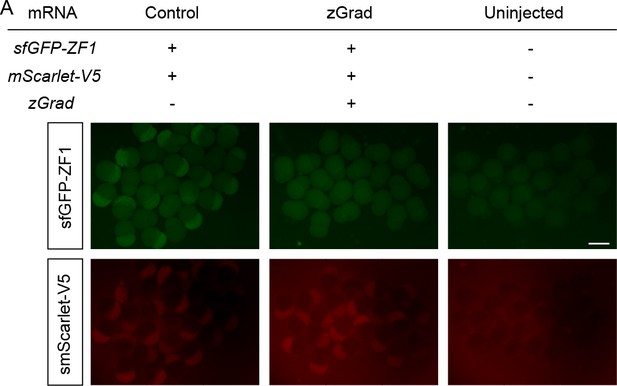
Onset of zGrad-mediated sfGFP-ZF1 degradation in early embryos.
(A) Images of embryos injected with sfGFP-ZF1 mRNA and mScarlet-V5 mRNA (left) or sfGFP-ZF1 mRNA and mScarlet-V5 mRNA and zGrad mRNA (middle) and uninjected control embryos (right) at 2.5 hpf. Scale bar: 1 mm.
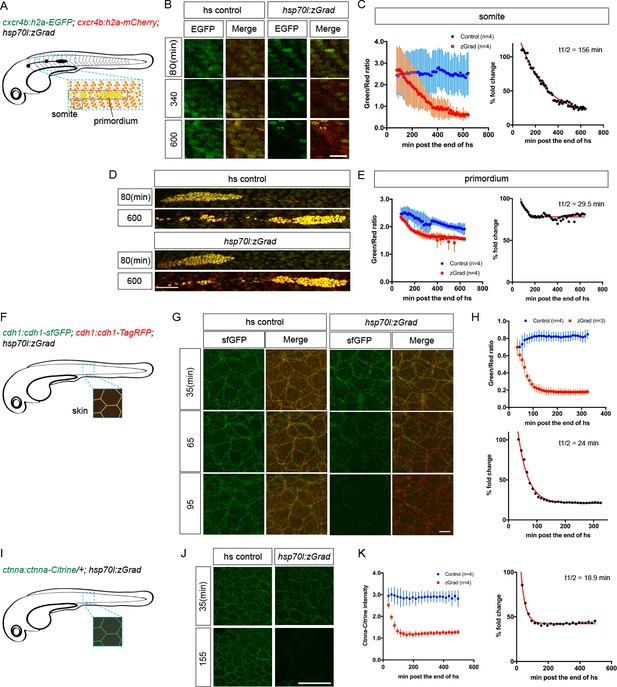
zGrad degrades nuclear, transmembrane and cytoplasmic proteins.
(A) Schematic of strategy to assess zGrad-mediated H2A-EGFP degradation in the somites and the primordium. (B) Maximum-projected confocal images of nuclei in the somites in heat-shocked cxcr4b:H2A-EGFP; cxcr4b:H2A-mCherry embryos transgenic for hsp70l:zGrad (right) or not (left) at indicated time in min after the end of heat shock (29–30 hpf). Scale bar: 20 µm. (C) Left, quantification of H2A-EGFP-to-H2A-mCherry ratios in the somites of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. Right, H2A-EGFP-to-H2A-mCherry ratio of zGrad-expressing embryos normalized to control embryos (black dots) and fitted to a one-exponential decay model (red). (D) Maximum-projected confocal images of primordium nuclei in heat-shocked cxcr4b:H2A-EGFP; cxcr4b:H2A-mCherry embryos transgenic for hsp70l:zGrad (bottom) or not (top) at indicated time in min after the end of heat shock (29–30 hpf). Scale bar: 50 µm. (E) Left, quantification of H2A-EGFP-to-H2A-mCherry ratios in the primordia of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. Right, H2A-EGFP-to-H2A-mCherry ratio of zGrad-expressing embryos normalized to control embryos (black dots) and fit to a one-exponential decay model (red). (F) Schematic of strategy to assess zGrad-mediated Cdh1-sfGFP degradation in the skin (enveloping and epidermal basal layer). (G) Maximum-projected confocal images of the skin in heat-shocked cdh1:cdh1-sfGFP; cdh1:cdh1-TagRFP embryos transgenic for hsp70l:zGrad (right) or not (left) at indicated time in min after the end of heat shock (31 hpf). Scale bar: 10 µm. (H) Top, quantification of Cdh1-sfGFP-to-Cdh1-TagRFP ratios in the primordia of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. Bottom, Cdh1-sfGFP-to-Cdh1-TagRFP ratio of zGrad-expressing embryos normalized to control embryos (black dots) and fit to a one-exponential decay model (red). (I) Schematic of strategy to assess zGrad-mediated Ctnna-Citrine degradation in the skin. (J) Maximum-projected confocal images of skin cells in heat-shocked ctnna:ctnna-Citrine/+ embryos transgenic for hsp70l:zGrad (right) or non-hsp70l:zGrad transgenic controls (left) at indicated time in min past the end of heat shock (31 hpf). Scale bar: 50 µm. (K) Left, quantification of Ctnna-Citrine levels in the skin of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. Right, Ctnna-Citrine levels in zGrad-expressing embryos normalized to Ctnna-Citrine levels in control embryos (black dots) and fit to a one-exponential decay model (red).
Degradation of H2A-EGFP by zGrad expressed from hsp70l promoter.
Heat-shocked hsp70l:zGrad; cxcr4b:h2a-EGFP; cxcr4b:h2a-mCherry embryo (top) and heat-shocked cxcr4b:h2a-EGFP; cxcr4b:h2a-mCherry control embryo (bottom). Time stamp indicates min after the end of the heat shock. Images are maximum-projected. Green: H2A-EGFP, Red: H2A-mCherry. Scale bar corresponds to 50 µm.
Degradation of Cdh1-sfGFP by zGrad expressed from hsp70l promoter.
Heat-shocked hsp70l:zGrad; cdh1:cdh1-sfGFP; cdh1:cdh1-TagRFP embryo (left) and heat-shocked cdh1:cdh1-sfGFP; cdh1:cdh1-TagRFP control embryo (right). Time stamp indicates min after the end of the heat shock. Images are maximum-projected. Green: Cdh1-sfGFP, Red: Cdh1-TagRFP. Scale bar corresponds to 10 µm.
Degradation of Ctnna-Citrine by zGrad expressed from hsp70l promoter.
Heat-shocked hsp70l:zGrad; ctnna:ctnna-Citrine/+ embryo (left) and heat-shocked ctnna:ctnna-Citrine/+ control embryo (Right). Time stamp indicates min after the end of the heat shock. Images are maximum-projected. Green: Ctnna-Citrine. Scale bar corresponds to 10 µm.
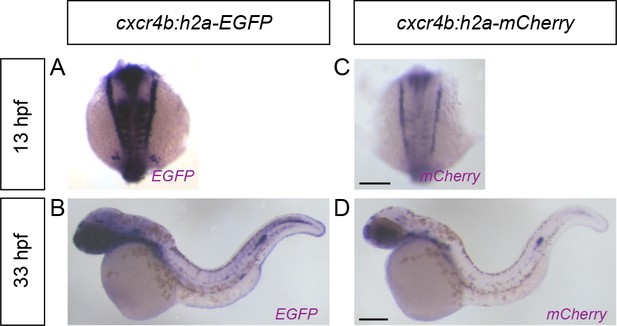
cxcr4b promoter activity during somitogenesis and primordium migration.
(A, B) Images of cxcr4b:h2a-EGFP embryos fixed at 13 hpf (A) and 33 hpf (B) and stained by in situ hybridization against EGFP mRNA. (C, D) Images of cxcr4b:h2a-mCherry embryos fixed at 13 hpf (C) and 33 hpf (D) and stained by in situ hybridization against mCherry mRNA. Scale Bars: 200 µm.
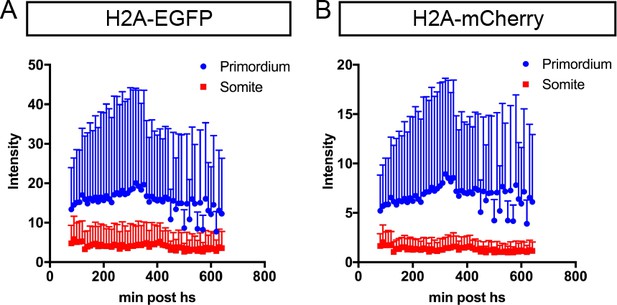
Fluorescent intensity of H2A-EGFP and H2A-mCherry in the somites and the primordium.
(A) Quantification of H2A-EGFP fluorescence intensity in the primordium (blue) and in the somites (red) of control embryos shown in Figure 2A–E. Time after the end of the heat shock is indicated in min on the x-axis. Mean and SD are indicated. Note that the H2A-EGFP fluorescence intensity in the primordium was on average 3.8 times brighter than the H2A-EGFP fluorescence intensity in the somites. (B) Quantification of H2A-mCherry fluorescence intensity in the primordium (blue) and in the somites (red) of control embryos in Figure 2A–E. Time after the end of the heat shock is indicated in min on the x-axis. Mean and SD are indicated. Note that the H2A-mCherry fluorescence intensity in the primordium was on average 5.1 times brighter than the H2A-mCherry fluorescence intensity in the somites.
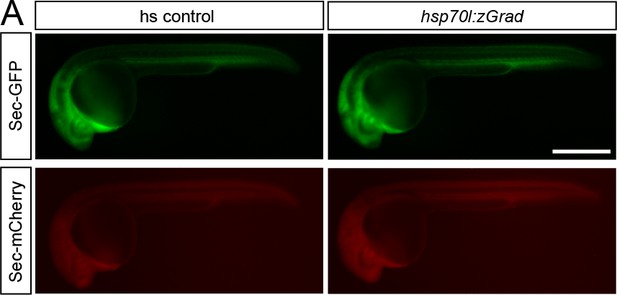
zGrad does not degrade secreted proteins.
(A) Images of hsp70l:sec-GFP; hsp70:sec-mCherry control embryos (left) and hsp70l:sec-GFP; hsp70:sec-mCherry; hsp70l:zGrad embryos (right) 5 hr post heat-shock at 30 hpf. Scale bar: 50 µm.
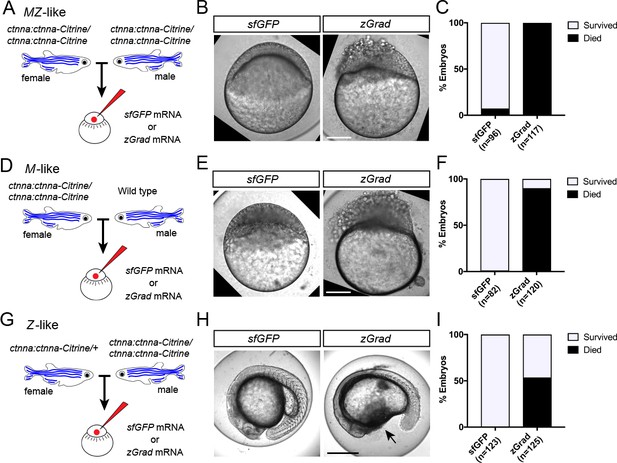
zGrad-mediated depletion of alpha-Catenin results in cell adhesion defects.
(A) Breeding strategy to assess zGrad-mediated degradation of maternally and zygotically provided Ctnna-Citrine on embryonic development. (B) Images of MZ ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA (left) or zGrad mRNA (right). Scale bar: 100 µm. (C) Quantification of MZ ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA or zGrad mRNA that disintegrated and died. (D) Breeding strategy to assess zGrad-mediated degradation of maternally provided Ctnna-Citrine on embryonic development. (E) Images of M ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA (left) or zGrad mRNA (right). Scale bar: 100 µm. (F) Quantification of M ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA or zGrad mRNA that disintegrated and died. (G) Breeding strategy to assess zGrad-mediated degradation of zygotically provided Ctnna-Citrine on embryonic development. (H) Images of Z ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA (left) or zGrad mRNA (right). Scale bar: 100 µm. (I) Quantification of Z ctnna:ctnna-Citrine embryos injected with sfGFP control mRNA and zGrad mRNA that displayed tissue rupture (arrow) and died.
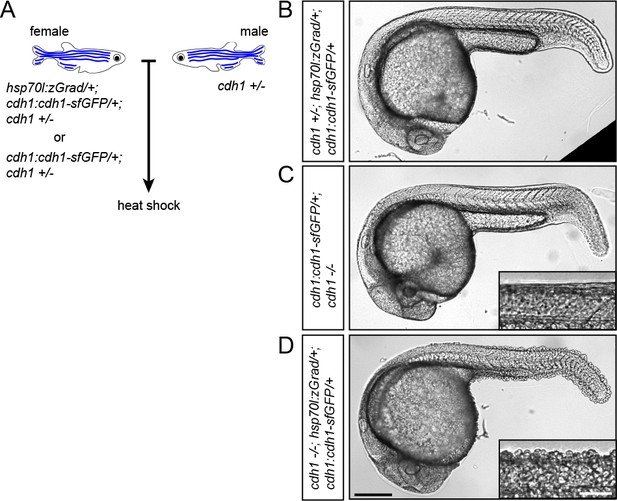
zGrad-mediated depletion of Cadherin-1 at 25 hpf results in skin defects and lethality.
(A) Breeding strategy to assess heat shock-induced zGrad-mediated degradation of Cdh1-sfGFP on embryonic development. (B, C, D) Images of cdh1+/-; hsp70l:zgrad/+; cdh1:cdh1-sfGFP/+ control embryo (B), cdh1-/-; cdh1:cdh1-sfGFP/+ control embryo (C) and cdh1-/-; hsp70l:zGrad/+; cdh1:cdh1-sfGFP embryo with skin defects (D) at 29 hpf. Embryos were heat shocked at 25 hpf for 1 hr. Scale bar: 200 µm. Insets in (C, D) are magnified images of the skin in the same embryos. Scale bar: 50 µm.
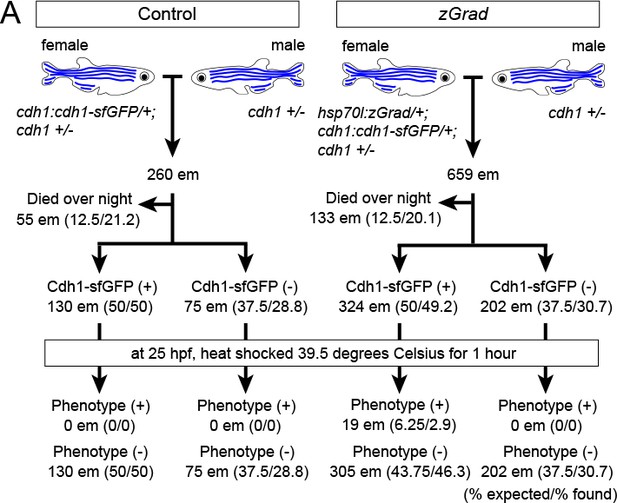
Expected results for zGrad-induced depletion of sfGFP-tagged Cadherin-1.
(A) Crossing scheme of indicated genotypes and % of embryos with expected and observed phenotype.
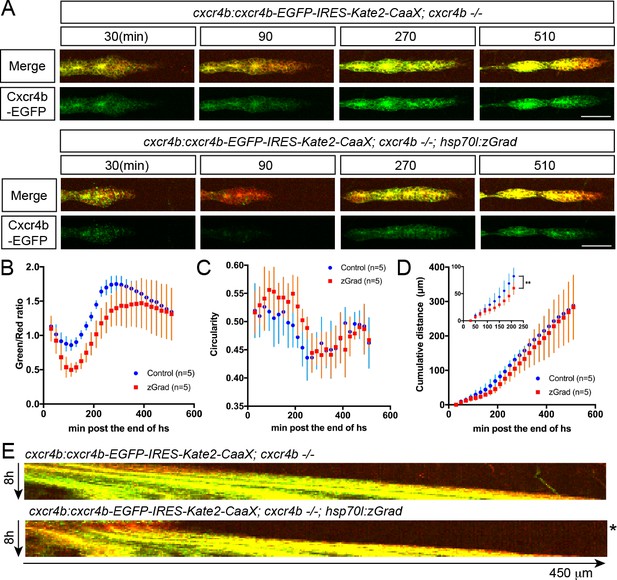
A pulse of zGrad degrades Cxcr4b-GFP and stalls primordium migration transiently.
(A) Maximum-projected confocal images of the primordia in heat-shocked cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p7; cxcr4b-/- embryos transgenic for hsp70l:zGrad (lower panels) or not (upper panels) at indicated time in min after the end of heat shock (31 hpf). Scale bar: 50 µm. (B) Quantification of Cxcr4b-EGFP-to-Kate2-CaaX ratios in the primordia of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. (C) Quantification of circularity of the primordia of control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. (D) Quantification of the cumulative primordium migration distance in control (blue) and zGrad-expressing embryos (red) after the end of heat shock in min. Mean and SD are indicated. Inset shows magnification of the 30 min to 240 min time interval. **=p < 0.01. (E) Kymograph of the primordia in heat-shocked cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p7; cxcr4b-/- embryos transgenic for hsp70l:zGrad (bottom) or not (top). Cxcr4b-EGFP is shown in green and Kate2-CaaX in red. Asterisk indicates the time interval in which Cxcr4b-EGFP is transiently degraded and the primordium transiently ceases to migrate.
Degradation of Cxcr4b-EGFP by zGrad expressed from hsp70l promoter.
Heat-shocked hsp70l:zGrad; cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p7; cxcr4b-/- embryo (top) and heat-shocked cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p7; cxcr4b-/- control embryo (Bottom). Time stamp indicates min after t he end of the heat shock. Images are maximum-projected. Green: Cxcr4b-EGFP, Red: Kate2-CaaX. Scale bar corresponds to 50 µm.
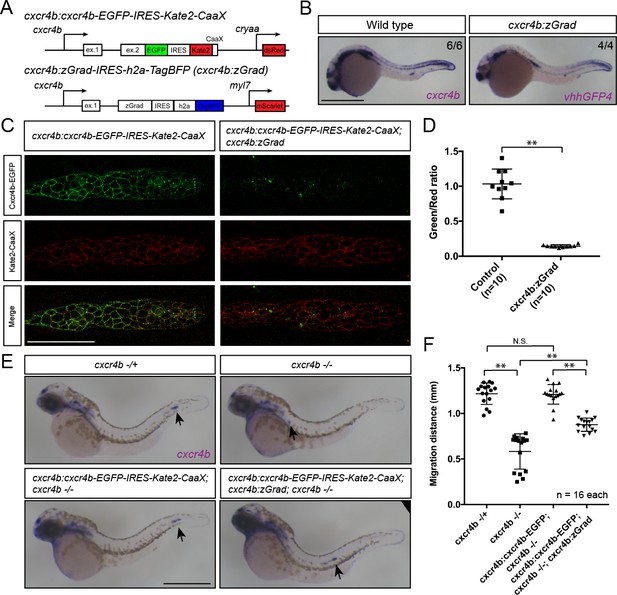
Tissue-specific expression of zGrad in the primordium degrades Cxcr4b-EGFP and slows down primordium migration.
(A) Schematic of strategy to assess zGrad-mediated Cxcr4b-EGFP degradation in the primordium on primordium migration. (B) In situ hybridization against cxcr4b mRNA in a wildtype embryo and against zGrad mRNA in a cxcr4b:zGrad embryo at 24hpf. Scale bar: 0.5 mm. (C) Single-plane confocal images of the primordium in cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p1 control (left) and cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p1; cxcr4b:zGrad embryos (right) at 36 hpf. Note that the embryos are cxcr4b +/- or cxcr4b -/-. Scale bar: 50 µm. (D) Quantification of Cxcr4b-EGFP to Kate2-CaaX fluorescence intensity ratio in the primordia of control embryos (blue) and embryos expressing zGrad in the primordium at 36 hpf. Mean and SD are indicated. **=p < 0.01. (E) In situ hybridization against cxcr4b mRNA in cxcr4b-/+ (top left), cxcr4b-/- (top right), cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p1; cxcr4b-/- (bottom left) and cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p1; cxcr4b-/-; cxcr4b:zGrad embryos (bottom right) at 38 hpf. Arrows indicate the location of the primordium. Scale bar: 0.5 mm. (F) Quantification of primordia migration distance of the indicated genotypes at 38 hpf. Mean, SD and n are indicated. **=p < 0.01, N.S. = p > 0.05.
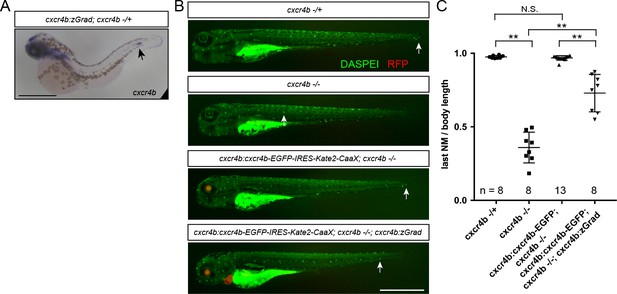
Tissue-specific Cxcr4b-EGFP degradation by zGrad.
(A) In situ hybridization against cxcr4b mRNA in a 38 hpf cxcr4b:zGrad; cxcr4b -/+ embryo. Arrow indicates the location of the primordium. Scale bar: 0.5 mm. (B) Images of live embryos of indicated genotypes stained for neuromasts with DASPEI at 4 dpf. Note that the cxcr4b:cxcr4b-EGFP-IRES-Kate2-CaaX-p1 transgene carries cryaa:dsRed, which expresses dsRED in the lens, and the cxcr4b:zGrad transgene carries myl7:mScarlet, which expresses mScarlet in the myocardium, as transgenic markers. Arrows indicate the position of the last neuromast. Scale bar: 1 mm. (C) Quantification of the position of the last neuromast normalized to body length of embryos shown in B. **=p < 0.01, N.S. = p > 0.05.
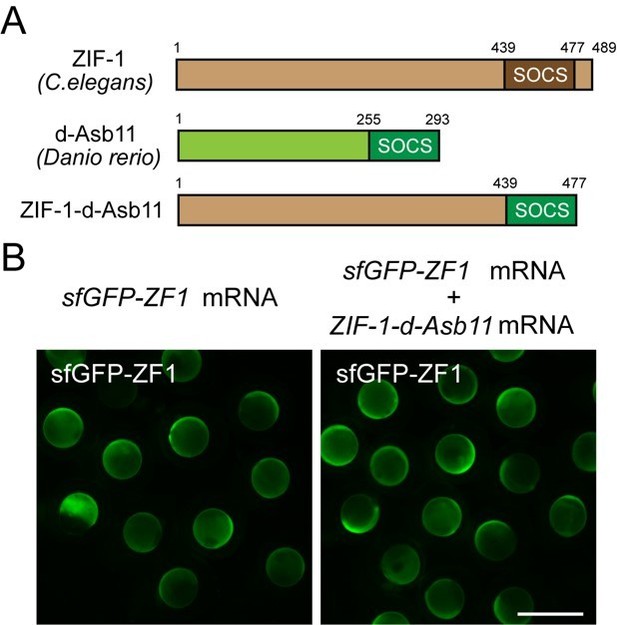
A zebrafish SOCS box does not increase ZIF-1-mediated degradation of ZF1-tagged GFP.
(A) Schematics of the chimeric protein design carrying ZF1-binding motif from ZIF-1 and SOCS box motif from d-Asb11. (B) Embryos injected with sfGFP-ZF1 mRNA or sfGFP-ZF1 and ZIF-1-d-Asb11 mRNA.
Tables
Cdh1 transgenic lines rescue cdh1 mutants.
https://doi.org/10.7554/eLife.43125.013| Transgenic line | Total number of embryos | Transgenic embryos phenotypically wild type | Non-transgenic embryos phenotypically wild type | Embryos phenotypically cdh1 mutant |
|---|---|---|---|---|
| cdh1:cdh1-sfGFP | 306 (100 %) | 149 (47 %) | 96 (33 %) | 61 (20 %) |
| cdh1:cdh1-TagRFP | 432 (100 %) | 204 (47 %) | 172 (40 %) | 56 (13%) |
Summary of degradation kinetics.
https://doi.org/10.7554/eLife.43125.014| FP-Tagged protein | Promoter expressing zGrad | Percent reduction | Degradation half-life in min | Time for onset of degradation in min |
|---|---|---|---|---|
| sfGFP-ZF1 | mRNA | 89% | N/A | N/A |
| Cxcr4b-EGFP | cxcr4b | 86% | N/A | N/A |
| H2A-EGFP (in somites) | hsp70l | 87% | 156 | 200 |
| H2A-EGFP (in primordium) | hsp70l | 22% | 29† | 140 |
| Cdh1-sfGFP | hsp70l | 79% | 24 | 75 |
| Ctnna-Citrine | hsp70l | 58% | 19 | 65 |
-
†Note that the zGrad-mediated degradation of H2A-EGFP in the primordium is obscured by the continued production of H2A-EGFP.
Additional files
-
Source code 1
ImageJ macro to analyze intensity of GFP and RFP channels of images from mRNA-injected embryos.
- https://doi.org/10.7554/eLife.43125.022
-
Source code 2
ImageJ macro to analyze signal intensity of the GFP and the RFP channel in time lapse movies from cxcr4b:h2a-EGFP; cxcr4b:h2a-mCherry embryos.
- https://doi.org/10.7554/eLife.43125.023
-
Source code 3
ImageJ macro to analyze signal intensity of the GFP and the RFP channel in time lapse movies from Cxcr4b-EGFP-to-Kate2-CaaX embryos.
- https://doi.org/10.7554/eLife.43125.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43125.025





