Cold-inducible RNA-binding protein (CIRBP) adjusts clock-gene expression and REM-sleep recovery following sleep deprivation
Figures
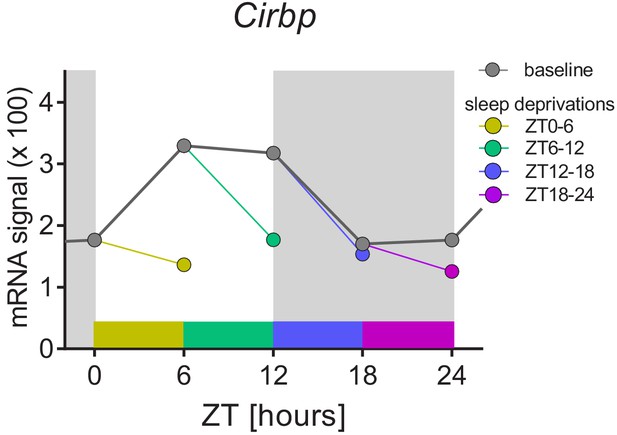
The sleep-wake distribution drives daily changes of Cirbp expression in the mouse brain.
Dark-grey symbols and line (baseline): from ZT0 to ZT12, mice spend most of their time asleep and Cirbp increases, whereas between ZT12-18, when mice spent most of their time awake, Cirbp decreases. When controlling for the daily occurrence in sleep by performing four 6 hr sleep deprivation starting at either ZT0, −6,–12, or −18 (each sleep deprivation is annotated with its own color), the diurnal amplitude of Cirbp is greatly reduced (colored circles represent level of Cirbp expression reached at the end of each sleep deprivation). Nine biological replicates per time point and condition were used (one data point missing at ZT18), and RNA was extracted from whole brain tissue (see Maret et al., 2007 for details). Data were taken from GEO GSE9442. Light-grey areas represent the dark periods.
-
Figure 1—source data 1
Expression of Cirbp in the brain under undisturbed conditions and after controlling for the circadian distribution of sleeping and waking.
- https://doi.org/10.7554/eLife.43400.003
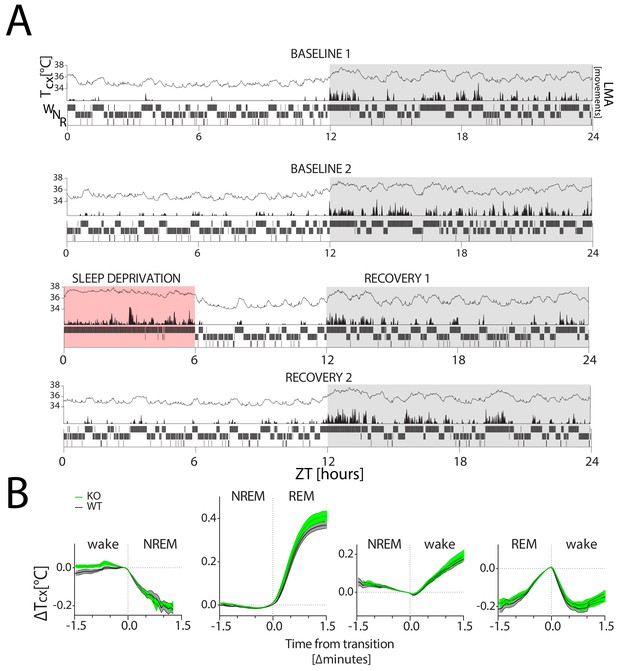
Tcx changes with sleep-wake state.
Tcx: cortical temperature; LMA: locomotor activity. (A) A representative 4-day recording of one mouse in LD 12:12 (in white:grey) during 2 baseline days (top two panels), followed by a 6 hr SD (in red; third panel) and 2 recovery days (bottom two panels), with within each panel Tcx (top; line graph), LMA (middle; area plot) and sleep-wake states (bottom; hypnogram). Sleep-wake states are averaged per minute to aid visualization. LMA was collected and plotted per minute (see Methods). (B) Changes in Tcx, depicted as mean ± SEM (WT: black lines, grey areas; KO: green lines and areas) relative to Tcx at the sleep-wake transition (average of the last value before and first value after transition). Tcx increased when transitioning from NREM sleep to wake and to REM sleep (two-way RM ANOVA, factors genotype (GT) and Time, factor Time, F(38,418)=126, F(45,540)=535.5; p<0.0001, respectively) and decreased when transitioning from wake to NREM sleep (F(22, 242)=1661, p<0.0001). Also the transition from REM sleep to wake affected the time course of Tcx (F(23,276)=131.8, p<0.0001). GT interacted with time during the wake to NREM sleep transition (F(22,242)=1.8, p=0.01), but not for other sleep-wake state transitions (p>0.12). No significant GT effect was detected (p>0.12) Transition data were obtained from both baseline days (see Materials and methods for details).
-
Figure 2—source data 1
Example of a 4-day recording; the transitions in sleep-wake state and associated changes in cortical temperature in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.005
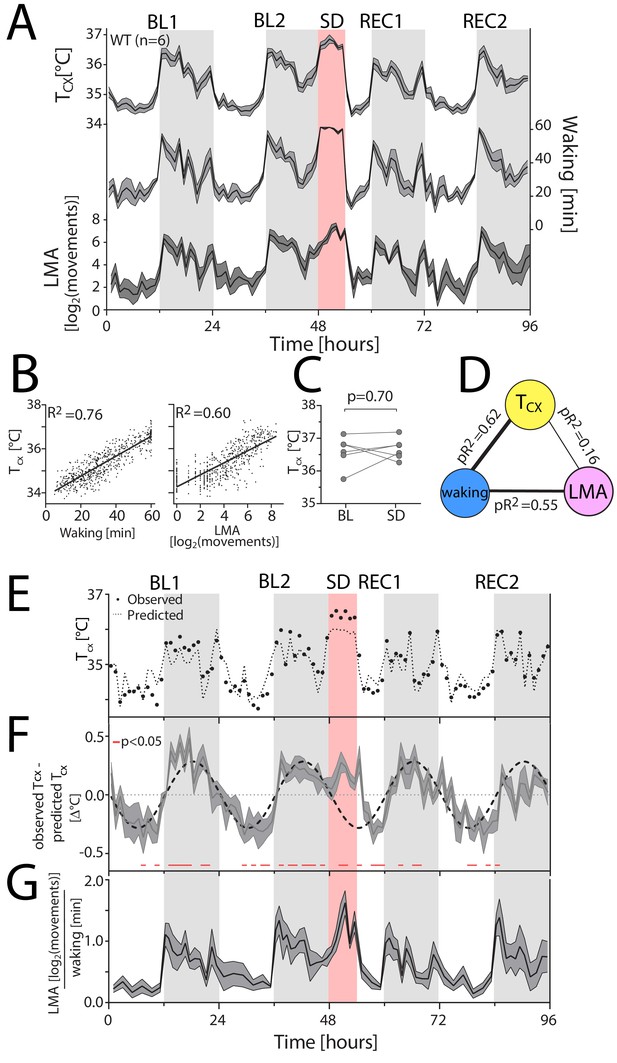
Waking is the major determinant of cortical temperature.
For panels A, F and G, the dark-grey line represents the mean, grey areas span ± 1 SEM. BL: baseline, SD: sleep deprivation, REC: recovery, Tcx: cortical temperature, LMA: locomotor activity. Light-grey areas mark the dark periods. (A) Time course of hourly values of Tcx, waking and LMA across the entire experiment. (B) Both waking (left) and LMA (right panel) strongly correlated with Tcx (n = 6; 96 values per mouse; p<0.0001). R2: correlation coefficients. (C) Tcx during SD did not differ from levels reached after long waking bouts during BL (t(5)=0.41, p=0.70). (D) Waking after correcting for LMA is the major determinant of Tcx, as revealed by partial correlation analysis; here performed on the combined hourly values of all WT mice. pR2: partial correlation coefficients. (E) A representative example [mouse TC03], with measured Tcx (closed circles), and predicted Tcx (stippled line) based on the correlation between Tcx and waking. (F) During the dark phase and SD, the predicted Tcx is lower than the measured Tcx, resulting in positive residuals [residuals: observed Tcx – predicted Tcx], whereas during the light phase, the predicted Tcx is higher than the measured Tcx, resulting in negative residuals (t-test: data <> 0, p<0.05, red lines underneath the curves). (G) LMA per unit of waking follows a similar pattern as the residuals in Panel F.
-
Figure 3—source data 1
Waking, LMA and cortical temperature.
- https://doi.org/10.7554/eLife.43400.011
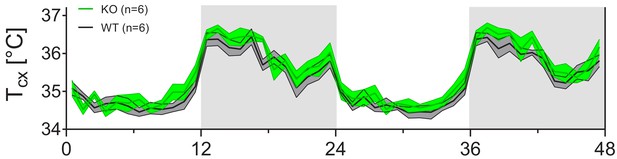
Cortical temperature (Tcx) shows similar daily variation in KO and WT mice.
The daily variation in Tcx does not differ between KO and WT mice (two-way RM ANOVA, factor Genotype (GT): F(1,5)=2.1, p=0.20; factor GT x Time: F(47,235)=0.93, p=0.61).
-
Figure 3—figure supplement 1—source data 1
Cortical temperature during baseline in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.008
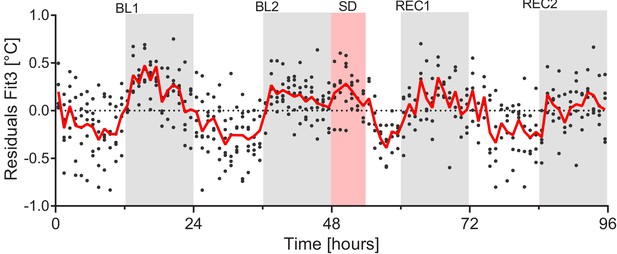
The residuals of the optimized mixed linear model.
Model3 (see Results section) still shows a pattern similar to that of residuals in Figure 3-F. Red line depicts the mean value determined every hour, each dot represents an hourly residual value per mouse.
-
Figure 3—figure supplement 2—source data 1
The residuals of the full model (LMA, Waking and LMA*Waking) explaining cortical temperature.
- https://doi.org/10.7554/eLife.43400.010
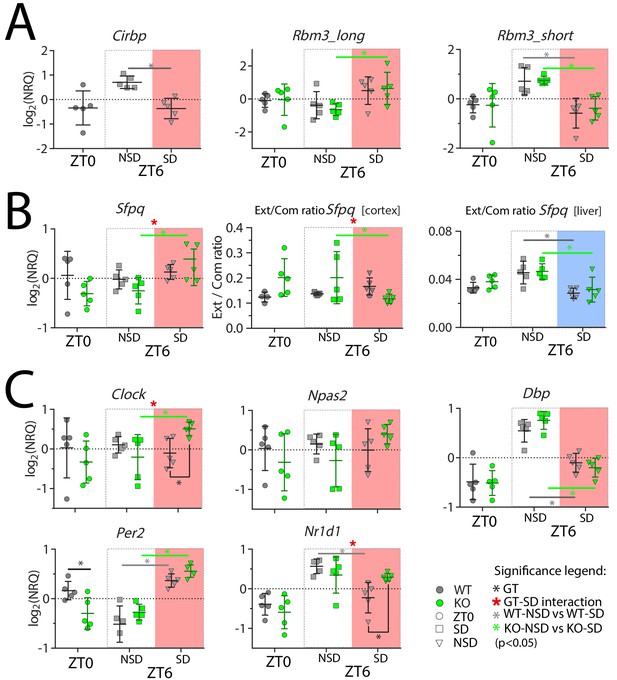
Cortical expression of several genes is affected by SD and by the lack of CIRBP.
NRQ: Normalized Relative Quantity, SD: sleep deprivation (salmon and blue areas for cortex and liver, respectively), NSD: non-sleep deprived (controls), GT: Genotype, ZT: Zeitgeber Time. n = 5 for each group, each symbol represents an observation in one mouse. Mice were sacrificed at ZT0, at ZT6 after sleep deprivation (ZT6-SD) or after sleeping ad lib (ZT6-NSD). Statistics are performed separately for ZT0 (factor GT, t-test), and ZT6 (factor GT and SD; two-way ANOVA). Significant (p<0.05) GT differences are indicated by a black line and *, the effect of SD in WT mice with a grey line and *, and in KO mice with a green line and *. GT-SD interactions at ZT6 are indicated by a red *. See Table 1 for statistics.
-
Figure 4—source data 1
Cortical expression of transcripts in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.018
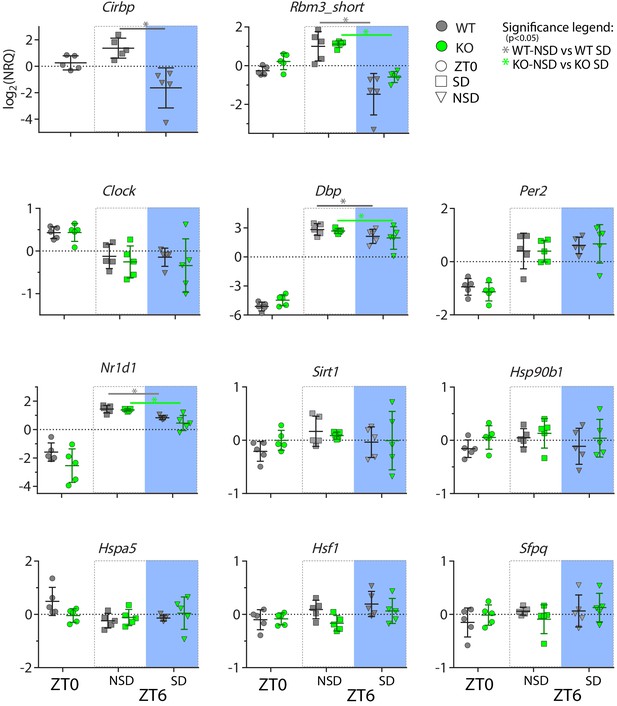
Changes in transcripts incurred by the absence of CIRBP and/or sleep deprivation in the liver.
SD: sleep deprivation, NSD: non sleep-deprived (controls), NRQ: normalized relative quantity, ZT: Zeitgeber Time, WT: wild type, KO: knock out. See Table 1 for statistics.
-
Figure 4—figure supplement 1—source data 1
Hepatic expression of transcripts in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.015
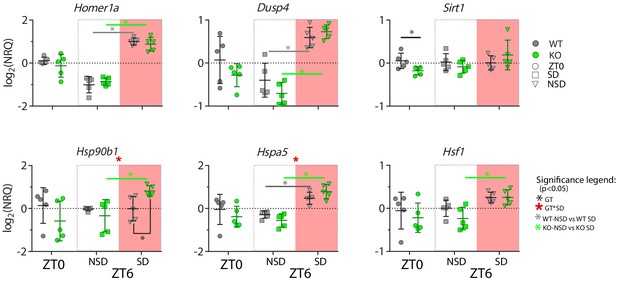
Changes in transcripts incurred by the absence of CIRBP and/or sleep deprivation in the cortex.
SD: sleep deprivation, NSD: non sleep-deprived (controls), NRQ: normalized relative quantity, ZT: Zeitgeber Time, WT: wild-type, KO: knock out. See Table 1 for statistics.
-
Figure 4—figure supplement 2—source data 1
Cortical expression of transcripts in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.017
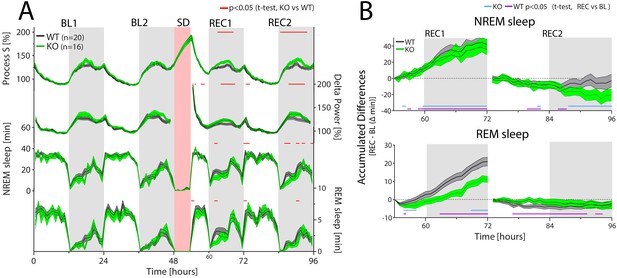
CIRBP modulates the sleep-wake distribution and REM sleep recovery after sleep deprivation.
Cirbp KO (green lines and areas) and WT (black line, grey areas) mice during the two baseline days (BL1 and −2), sleep deprivation (SD), and the 2 recovery days (REC1 and −2; areas span ± 1 SEM range). (A) From top to bottom: Simulated delta power (Process S), measured NREM sleep delta power, NREM and REM sleep. Only during REC, both Process S and delta power are increased in Cirbp KO mice compared to WT (F(1,34)=5.56, p=0.024; F(1,34)=4.65, p=0.038, respectively), based on differences in NREM-sleep distribution (Genotype (GT): F(1,34)=6.02, p=0.0194). GT effects in REM sleep were also detected during recovery (factor GT: F(1,34)=5.45, p=0.026). Exact timing of GT differences is indicated by red lines above each graph (post-hoc t-test, p<0.05). (B) Top: KO mice recover as much NREM sleep as WT mice do in the first 18 hr after SD (REC1 at 72 hr: WT: 41.9 ± 6.1 KO: 38.6 ± 9.7 min; t-test: t(34)=0.30, p=0.76). Bottom: KO mice accumulated less REM sleep during the first recovery day over the baseline day in comparison to WT mice relative to baseline (REC1 at 72 hr, WT: 20.9 ± 2.3 KO: 9.9 ± 2.0, t-test: t(34)=3.7, p=0.0007). Recovery-to-baseline differences are indicated by blue (KO) and purple (WT) lines below each graph (post-hoc t-test, p<0.05). Light-grey areas mark the dark periods.
-
Figure 5—source data 1
Simulated Process S, delta power, NREM and REM sleep in Cirbp WT and KO mice during two baseline days, a 6hr sleep deprivation and two recovery days.
- https://doi.org/10.7554/eLife.43400.021
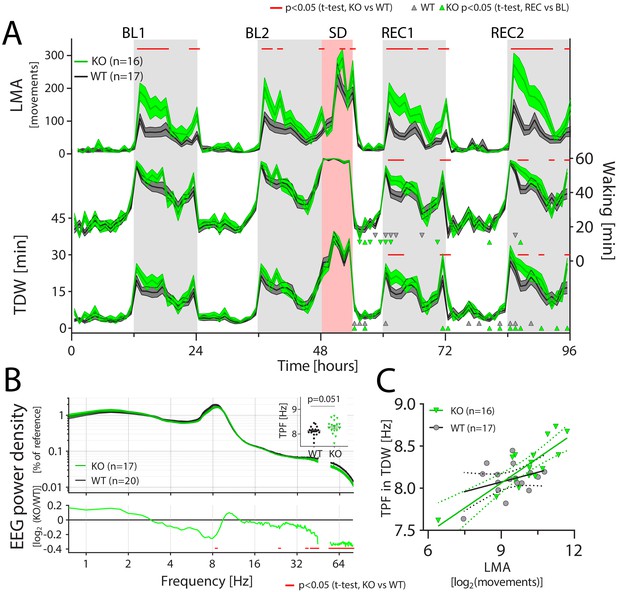
CIRBP suppresses locomotor activity and affects spectral composition during theta-dominated waking.
LMA: locomotor activity, TDW: theta-dominated waking, TPF: theta-peak frequency, GT: genotype. (A) Cirbp KO (green lines and areas) and WT (black line, dark-grey areas) mice during the two baseline days (BL1 and −2), sleep deprivation (SD), and the two recovery days (REC1 and −2; areas span ± 1 SEM range). Cirbp KO mice are more active in the dark periods (light-grey areas) only (BL: GTxTime: F (47,1457)=3.5, p<0.0001; REC: GTxTime: F (41,1271)=5.2, p<0.001), and spent more time awake and inTDW during REC compared to WT mice (total waking: BL: GTxTime: F(47,1457) = 1.1, p=0.33 REC: GTxTime: F (41 1271)=1.9, p=0.0005; TDW: BL: GTxTime: F (47,1457)=1.1, p=0.35; REC: GTxTime: F (41,1271)=1.8, p=0.0025). Significant genotype differences are marked by red lines above each graph (post-hoc t-tests; p<0.05). ∆ and ∇ indicate a significant increase and decrease in REC compared to same time in BL, respectively. (B) CIRBP contributes to the spectral composition of TDW in the dark phase (two-way RM ANOVA; GTxFreq: F(278,9730) = 2.0; p<0.0001, red symbols in lower panel: post-hoc t-tests, p<0.05), and KO mice tend to have faster TPF during TDW in the dark phase (t(35)=2.0; p=0.0506). (C) TPF in the dark phase correlates only in the KO mice significantly with LMA (WT: R2 = 0.12, p=0.17, KO: R2 = 0.71, p<0.0001).
-
Figure 6—source data 1
Time course of LMA, waking and theta-dominated waking in Cirbp WT and KO mice; spectral composition of theta-dominated waking, and relation between theta-peak frequency in theta-dominated waking and LMA.
- https://doi.org/10.7554/eLife.43400.027
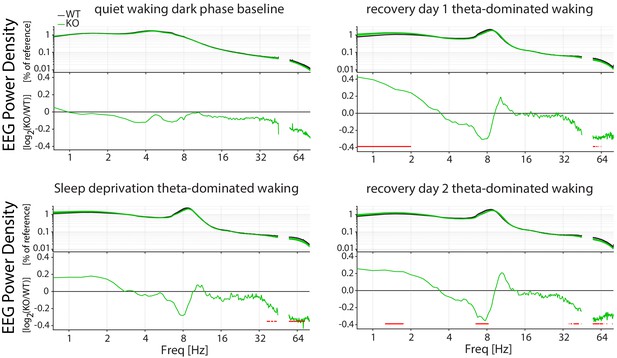
Changes in the EEG spectra are observed in theta-dominated waking (TDW), but not in quiet waking.
Theta-dominated waking: TDW, Genotype: GT. Spectral composition of Cirbp KO (green lines and areas; n = 17) and WT (black line, grey areas; n = 20) mice at different times of the experiment (indicated by the title, BL: baseline, REC1: recovery1, REC2: recovery2). In each graph, the top panel depicts the EEG spectral composition, whereas in the lower panel the KO spectral composition relative to the WT is shown (ratio KO/WT). Red symbols in lower panel indicate significant difference for the frequency bins (post-hoc t-test, p<0.05). ‘Quiet’ waking baseline dark phase: No effect of GT or an interaction effect (two-way RM ANOVA, GT*Frequency: F(278, 9730)=0.81, p=0.99) on the spectral composition of quit waking in the dark phase of the baseline. Sleep deprivation TDW: significant interaction between GT and Frequency (F (278, 9730)=1.722, p<0.0001). REC1 TDW: significant interaction: F (278, 9730)=2.984, p<0.0001; REC2 TDW: significant interaction F(278, 9730)=3.083, p<0.0001).
-
Figure 6—figure supplement 1—source data 1
Spectral composition of the waking EEG in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.024
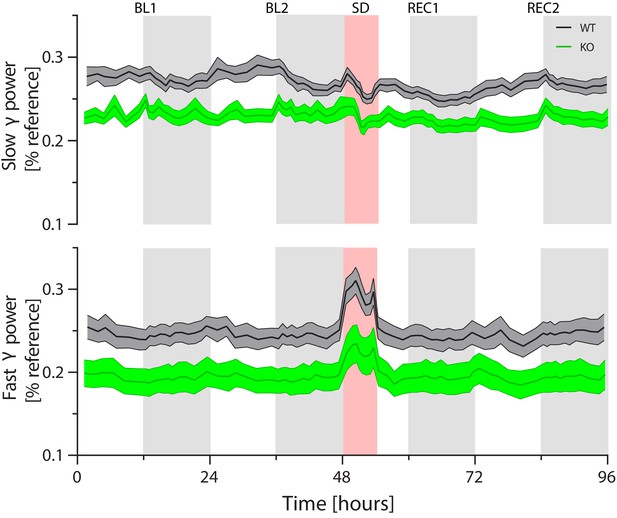
Slow and fast gamma power over the course of the experiment in theta-dominated waking.
Cirbp KO (green lines and areas) and WT (black line, grey areas) mice (areas span ± 1 SEM range). BL: baseline, SD: Sleep deprivation, REC: Recovery, Genotype; GT. Power in both slow [32–45 Hz] and fast [55–80 Hz] gamma is significantly reduced over the course of the experiment in Cirbp KO mice (two-way ANOVA: factor GT: slow gamma: F(1,34)=11.9, p=0.002; fast gamma: F(1,33)=6.6, p=0.01). Like the analysis of delta power, power in the gamma bands is calculated based on intervals to ensure an equal contribution of epochs to each time point: six intervals in the light phase, 12 in the dark periods (light-grey areas), eight in the sleep deprivation and four during the recovery light period.
-
Figure 6—figure supplement 2—source data 1
Time course of fast and slow gamma power during theta-dominated waking in Cirbp WT and KO mice.
- https://doi.org/10.7554/eLife.43400.026
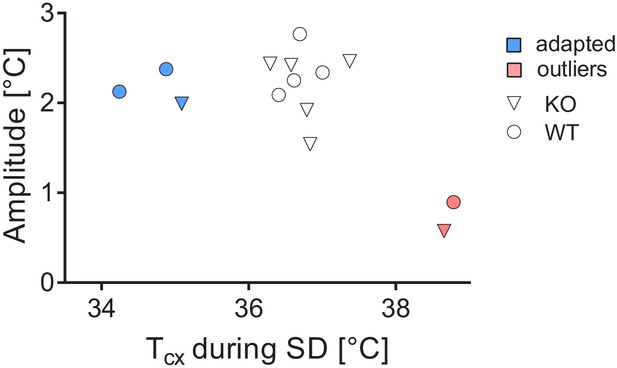
Assessment of amplitude and absolute values of cortical temperatures.
Two outliers were detected (pink), whereas three others were corrected for their low values (blue).
-
Figure 7—source data 1
The daily amplitude of cortical temperature and cortical temperature reached during sleep deprivation.
- https://doi.org/10.7554/eLife.43400.032
Tables
Statistics on RT-qPCR results.
https://doi.org/10.7554/eLife.43400.012| Cortex | Liver | |||||||
|---|---|---|---|---|---|---|---|---|
| ZT6 | ZT6 | |||||||
| Transcript | ZT0 | SD/NSD | GT | Interaction | ZT0 | SD/NSD | GT | Interaction |
| Cirbp | X | X | X | X | X | X | ||
| Clock | t = 0.86; p=0.21 | F = 2.38; p=0.14 | F = 0.85; p=0.37 | t = 0.03; p=0.98 | F = 0.09; p=0.77 | F = 0.81; p=0.38 | F = 0.03; p=0.87 | |
| Dbp | t = 0.13; p=0.90 | F = 0.39; p=0.54 | F = 3.06; p=0.10 | t = 1.99; p=0.08 | F = 0.23; p=0.64 | F = 0.0002; p=0.99 | ||
| Dusp4 | t = 1.29;p=0.23 | F = 0.50; p=0.49 | F = 3.24; p=0.09 | X | X | X | X | |
| Homer1a | t = 0.96; p=0.36 | F = 0.005; p=0.94 | F = 1.08; p=0.31 | X | X | X | X | |
| Hsf1 | t = 0.67;p=0.52 | F = 1.79; p=0.20 | F = 1.9; p=0.18 | t = 0.14; p=0.89 | F = 3.43; p=0.08 | F = 4.63; p=0.05 | F = 0.48; p=0.50 | |
| Hsp90b1 | t = 1.29; p=0.23 | F = 1.40; p=0.25 | t = 1.71; p=0.12 | F = 0.93; p=0.35 | F = 0.80; p=0.38 | F = 0.07; p=0.80 | ||
| Hspa5 | t = 0.89; p=0.40 | F = 0.03; p=0.86 | t = 2.02; p=0.08 | F = 0.62; p=0.44 | F = 0.84; p=0.37 | F = 0.04; p=0.86 | ||
| Npas2 | t = 0.86; p=0.41 | F = 1.56; p=0.2298 | F = 0.0008;p=0.98 | F = 3.99; p=0.06 | X | X | X | X |
| Per2 | F = 0.06; p=0.80 | t = 0.90; p=0.40 | F = 0.95; p=0.34 | F = 0.01; p=0.92 | F = 0.02; p=0.90 | |||
| Rbm3-short | t = 0.05 ; p=0.96 | F = 0.31; p=0.59 | F = 0.13; p=0.73 | t = 2.23; p=0.06 | F = 2.7; p=0.12 | F = 1.6; p=0.22 | ||
| Rbm3-long | t = 0.10 ; p=0.92 | F = 0.03; p=0.86 | F = 0.32; p=0.58 | X | X | X | X | |
| Nr1d1 | t = 0.91; p=0.39 | F = 1.09; p=0.31 | t = 1.59; p=0.15 | F = 2.41; p=0.14 | F = 1.37; p=0.26 | |||
| Sfpq | t = 1.51; p=0.17 | F = 0.017; p=0.90 | t = 0.93; p=0.38 | F = 1.26; p=0.28 | F = 2.78; p=0.11 | F < 0.001;p=0.98 | ||
| Sirt1 | F = 1.61;p=0.22 | F = 0.14; p=0.72 | F = 2.07; p=0.17 | t = 1.75; p=0.12 | F = 0.94; p=0.35 | F = 0.03; p=0.87 | F = 0.12; p=0.73 | |
-
GT: genotype, SD/NSD: Sleep deprived / non-sleep deprived (control).
ZT0: t-test, degrees of freedom (df): 8.
-
ZT6: two-way ANOVA (factors SD and GT), df = 1 for both factors SD, GT and its interaction; error df = 16.
X: Ct >30 or undetected.
-
: significant decrease (at ZT0: KO relative to WT; at ZT6, SD/NSD: SD relative to NSD; GT: KO relative to WT).
: significant increase (at ZT0: KO relative to WT; at ZT6, SD/NSD: SD relative to NSD; GT: KO relative to WT).
-
: significant interaction. Significance level: α = 0.05.
Baseline time spent in sleep-wake states, including theta-dominated waking (TDW; min), and locomotor activity (LMA; movements) per 12 hr per genotype (mean ±1 SEM), averages of BL1-2.
Two-way ANOVA (Factor GT and Light/Dark) on those same 12 hr values. Degrees of freedom for both GT and Light/Dark: df = 1; error term: df = 35.
| WT | KO | Statistics (Two-way ANOVA) | |||
|---|---|---|---|---|---|
| Light | Dark | Light | Dark | Factor GT x Light/Dark, df : 1,35 | |
| NREM sleep | 389 ± 4 | 189 ± 10 | 376 ± 4 | 170 ± 13 | F = 0.02, p=0.89 |
| REM sleep | 70 ± 2 | 19 ± 2 | 66 ± 2 | 20 ± 2 | F = 0.83, p=0.37 |
| Total waking | 260 ± 4 | 512 ± 11 | 277 ± 5 | 530 ± 14 | F = 0.02, p=0.90 |
| TDW | 45 ± 3 | 179 ± 12 | 55 ± 5 | 192 ± 15 | F = 0.13, p=0.72 |
| LMA | 119 ± 16 | 817 ± 70 | 181 ± 26 | 1370 ± 142 | F = 7.1, p=0.01 |
Time constants, asymptotes and So for Process S do not differ between Cirbp WT and KO mice.
Mean time constants (±SEM) obtained by the simulation (Process S) with the best fit to the NREM sleep delta power values, where the increase of Process S is simulated with the time constant τi, the decrease with τd and the upper- and lower asymptotes by UA and LA, respectively. S0 is the level of Process S at time = 0. No significant genotype differences were observed. See Material and methods for detailed description of the simulation. Degrees of freedom: df.
| WT | KO | t-test, df = 34 | |
|---|---|---|---|
| S0 [%] | 128.2 ± 2.4 | 132.1 ± 2.6 | t = 1.10, p=0.29 |
| τi [h] | 13.2 ± 1.2 | 12.9 ± 1.0 | t = −0.16, p=0.87 |
| τd [h] | 3.0 ± 0.2 | 2.8 ± 0.2 | t = −0.74, p=0.46 |
| LA [%] | 45.1 ± 1.4 | 45.1 ± 1.1 | t = −0.02, p=0.98 |
| UA [%] | 288.8 ± 3.0 | 296.6 ± 3.2 | t = 1.80, p=0.09 |
Theta-peak frequency (TPF; mean ±SEM in [Hz]) during theta-dominated waking (TDW) in the baseline light and dark periods.
https://doi.org/10.7554/eLife.43400.029| TPF during BL | WT | KO |
|---|---|---|
| Light | 7.77 ± 0.03 | 7.64 ± 0.04 |
| Dark | 8.13 ± 0.04 | 8.28 ± 0.07 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. Musculus (male)) | Cirbp KO; Cirbp WT | PMID: 22711815 | RRID:MGI:5432528 | Professor Jun Fujita (Kyoto University) |
| Sequence-based reagent | RT-qCPR primers | This paper. | See Table 5 | |
| Commercial assay or kit | RNeasy Lipid Tissue Mini Kit 50 | Qiagen | Catalog no. 74804 | |
| Commercial assay or kit | RNeasy Plus Mini Kit 50 | Qiagen | Catalog no. 74134 | |
| Commercial assay or kit | Invitrogen Superscript II reverse transcriptase | Thermo Fisher | Catalog no. 18064022 | |
| Commercial assay or kit | TaqMan mastermix | Thermo Fisher | Catalog no. 4369510 |
Sequences of the forward and reverse primer and probe used for the RT-qPCR.
https://doi.org/10.7554/eLife.43400.030| GeneName | FwdPrimer | RevPrimer | Probe |
|---|---|---|---|
| Cirbp | AGGGTTCTCCAGAGGAGGAG | CCGGCTGGCATAGTAGTCTC | CGCTTTGAGTCCCGGAGTGGG |
| Clock | CGAGAAAGATGGACAAGTCTACTG | TCCAGTCCTGTCGAATCTCA | TGCGCAAACATAAAGAGACCACTGCA |
| Dbp | CGTGGAGGTGCTTAATGACCTTT | CATGGCCTGGAATGCTTGA | AACCTGATCCCGCTGATCTCGC |
| Dusp4 | GTTCATGGAAGCCATCGAGT | CCGCTTCTTCATCATCAGGT | TCCCGATCAGCCACCATCTGC |
| Eef1a2 | CCTGGCAAGCCCATGTGT | TCATGTCACGAACAGCAAAGC | TGAGAGCTTCTCTGACTACCCTCCACTTGGT |
| Gadph | TCCATGACAACTTTGGCATTG | CAGTCTTCTGGGTGGCAGTGA | AAGGGCTCATGACCACAGTCCATGC |
| Homer1a | GCATTGCCATTTCCACATAGG | ATGAACTTCCATATTTATCCACCTTACTT | ACA5ATT5AATT5AG5AATCATGA (*) |
| Hsf1 | CAACAACATGGCTAGCTTCG | CTCGGTGTCATCTCTCTCAGG | TGAGCAGGGTGGCCTGGTCA |
| Hsp90b1 | TGTACCCACATCTGCACCTC | TTGGGCATCATATCATGGAA | CGCCGCGTATTCATCACAGATGA |
| Hspa5 | CACTTGGAATGACCCTTCG | GTTTGCCCACCTCCAATATC | TGGCAAGAACTTGATGTCCTGCTGC |
| Npas2 | AGGAAAGGACGTCTGCTTCA | CCAAGCTATGCCTCGAAGTG | CCTGGCAACCCCGCAGTTCTTA |
| Per2 | ATGCTCGCCATCCACAAGA | GCGGAATCGAATGGGAGAAT | ATCCTACAGGCCGGTGGACAGCC |
| Rbm3-long | TGATGCTGTCTTCAGGATGC | GGCCCAACACAAGTAAAGGA | TCAAGGATGAGGTAAGTATGCTATCCTTGAGC |
| Rbm3-short | GGCTATGACCGCTACTCAGG | CAGCAATTTGCAAGGACGAT | TGAGATGGGGCATGCACACA |
| Nr1d1 | AGGGCACAAGCAACATTACC | CAGGCGTGCACTCCATAGT | AGGCCACGTCCCCACACACC |
| Sfpq | GCATTTGAAAGATGCAGTGAA | CAGGAAGACCATCTTCGTCA | TCGCCCAGTCATTGTGGAACCA |
| Sfpq_Comm | TGGATGTTAGCAGTTTATTGACC | GCACAAGGTACACTGCCATT | TGTAAATGGCCTGTTTGGGCAGG |
| Sfpq_Ext | TGCTTTCCTCCCACCATAAG | TTGCTCTAACGAAAGGAAATTCA | TGGGGATGTTTTGATGATGTCAGTTCA |
| Sirt1 | TTGTGAAGCTGTTCGTGGAG | CTCATCAGCTGGGCACCTA | TTTTAATCAGGTAGTTCCTCGGTGCCC |
| Tbp | TTGACCTAAAGACCATTGCACTTC | TTCTCATGATGACTGCAGCAAA | TGCAAGAAATGCTGAATATAATCCCAAGCG |
-
(*) 5 = propynyl dC; increases the melting temperature of the probe.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43400.033





