Temporal identity establishes columnar neuron morphology, connectivity, and function in a Drosophila navigation circuit
Figures
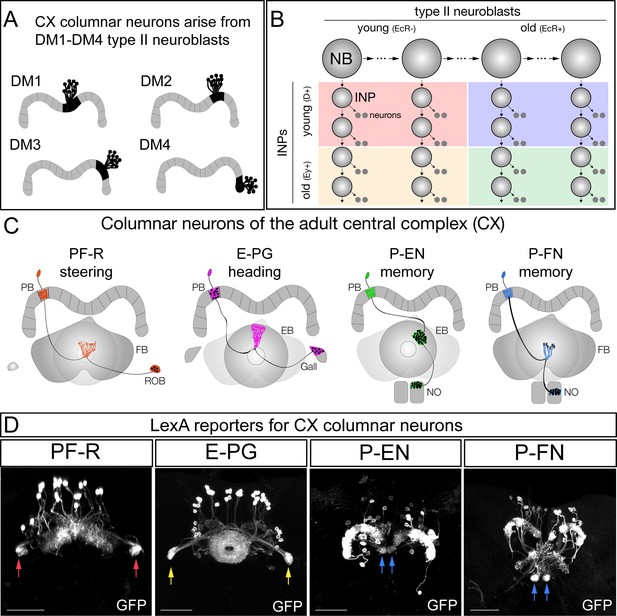
CX columnar neurons are generated by type II neuroblast lineages.
(A) CX columnar neurons innervating the PB originate specifically from each of four bilateral type II neuroblast lineages (DM1-DM4), which include all four neuronal subtypes shown in panel D. DM1 lineage neurons innervate the most medial PB glomeruli, and DM4 lineage neurons innervate the most lateral PB glomeruli. Adult brain right hemisphere shown. (B) Type II neuroblasts divide every 1.6 hr to generate ~60 INPs; each INP progeny divides every 2–3 hr to produce 10–12 neurons (Homem et al., 2013). Both neuroblasts and INPs express temporal transcription factors that subdivide their lineages into distinct molecular windows. Finer subdivisions exist but are not shown for clarity. (C) PF-R, E-PG, P-EN, and P-FN columnar neuron subtypes; each has a proposed function in navigation (Stone et al., 2017; ) and a distinct pattern of connectivity. PB, protocerebral bridge; FB, fan-shaped body; ROB, round body; EB, ellipsoid body; NO, noduli. (D) Adult CX columnar neurons derived from INPs labeled with adult LexA lines specific for each subtype; see Figure 1—figure supplement 1A for genetic details. ROB, red arrows; Gall, yellow arrows; Noduli, blue arrows. Scale bars, 40µm. Genotypes: PF-R, UAS-FLP; R9D11-Gal4, R37G12-lexA; lexAop(FRT.stop)mCD8::GFP; E-PG, UAS-FLP; R9D11-Gal4, R60D05-lexA; lexAop(FRT.stop)mCD8::GFP; P-EN, UAS-FLP; R9D11-Gal4, R12D09-lexA; lexAop(FRT.stop)mCD8::GFP; P-FN, UAS-FLP; R9D11-Gal4, R16D01-lexA; lexAop(FRT.stop)mCD8::GFP.
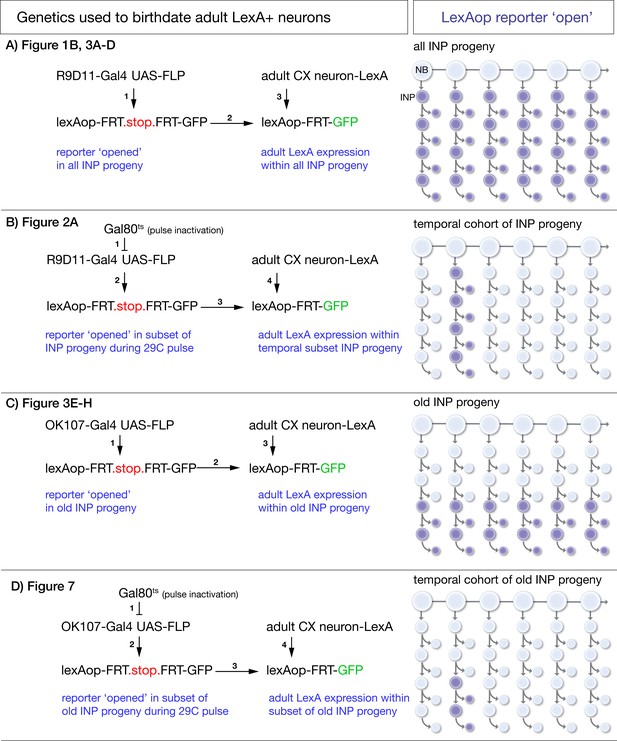
Intersectional genetic birthdating schemes.
(A) Identifying columnar neuron subtypes derived from all INPs. R9D11-Gal4 drives expression of FLP which excises an FRT-stop in all INPs and their progeny. This allows columnar neuron specific LexA lines to drive GFP expression if the neurons derive from INPs. (B) Identifying the time during the neuroblast lineage that produces each columnar neuron subtype. A pulse of 29°C disables ts.Gal80 to allow R9D11-Gal4 to excise FRT.stop in the INPs present during the heat pulse. This allows columnar neuron specific LexA lines to drive GFP expression if the neurons derive from INPs born from the type II neuroblast at the time of heat pulse. (C) Identifying columnar neuron subtypes derived from young or old INPs. OK107-Gal4 drives expression of FLP which excises an FRT-stop only in old INPs and their progeny. This allows columnar neuron specific LexA lines to drive GFP expression if the neurons derive from old INPs. Lack of expression shows the neurons are derived from young INPs. (D) Identifying the time during the neuroblast lineage that produces each old INP-derived columnar neuron subtype. A pulse of 29°C disables ts.Gal80 to allow OK107-Gal4 to excise the FRT.stop in the INPs present during the heat pulse. This allows columnar neuron specific LexA lines to drive GFP expression if the neurons derive from INPs present at the time of heat pulse.
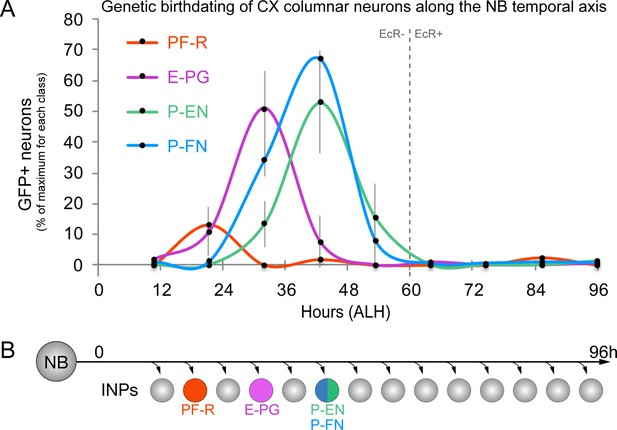
CX columnar neurons are generated by young type II neuroblast lineages.
(A) Identifying the time during the neuroblast lineage that produces each columnar neuron subtype. See Figure 1—figure supplement 1B for genetic details. Note that PF-R neurons are born first, E-PG neurons second, and then P-EN/P-FN neurons sharing a common birthdate (n = 3–6 per time-point). (B) Summary of NB birthdating results.
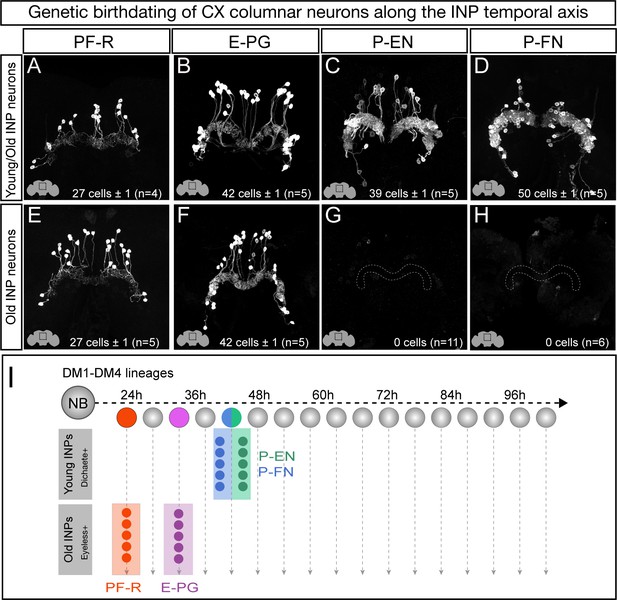
Each CX columnar neuron type arises exclusively from young or old INP lineages.
(A–D) Columnar neuron cell bodies labeled by subtype-specific LexA lines derive from INP lineages (n = 5 for each experiment). Staining shows the volume containing cell bodies; thus most axon and dendrite projections are not visible. See Figure 1—figure supplement 1A for genetic details. (E–H) The PF-R and E-PG columnar neurons are generated by late INPs (n = 5 for each experiment), whereas the P-EN (n = 11) or P-FN (n = 6) neurons were not derived from old INPs and thus are fully derived from young INPs. Staining shows the volume containing cell bodies; thus most axon and dendrite projections are not visible. See Figure 1—figure supplement 1C for genetic details. (I) Summary of INP birthdating results.
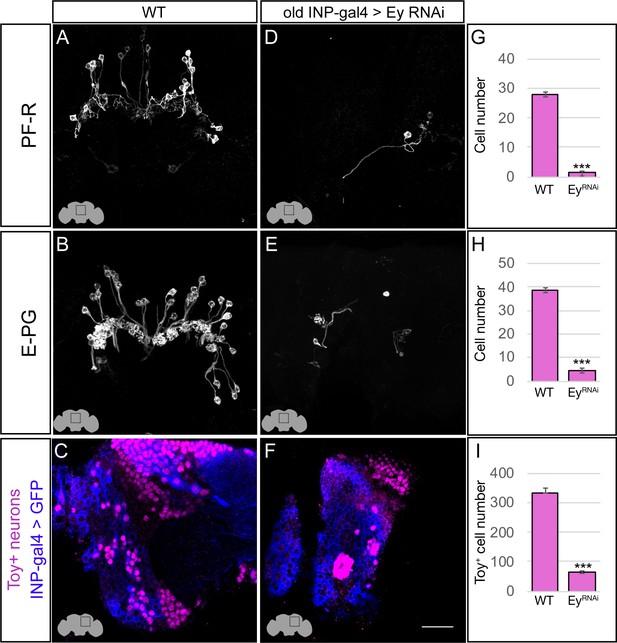
Eyeless promotes PF-R and E-PG molecular identity.
(A–C) Wild-type PF-R, E-PG, and Toy+ neurons in the dorsoposterior adult brain. PF-R and E-PG neurons detected by expression of neuron-specific LexA lines. See Materials and methods for genotypes. (D–F) EyelessRNAi in INP lineages decreases the number of PF-R, E-PG, and Toy+ late-born neurons in the dorsoposterior adult brain. See Materials and methods for genotypes. (G–I) Quantification (n = 5 for each experiment). ***, p<0.001. Scale bar, 20µm.
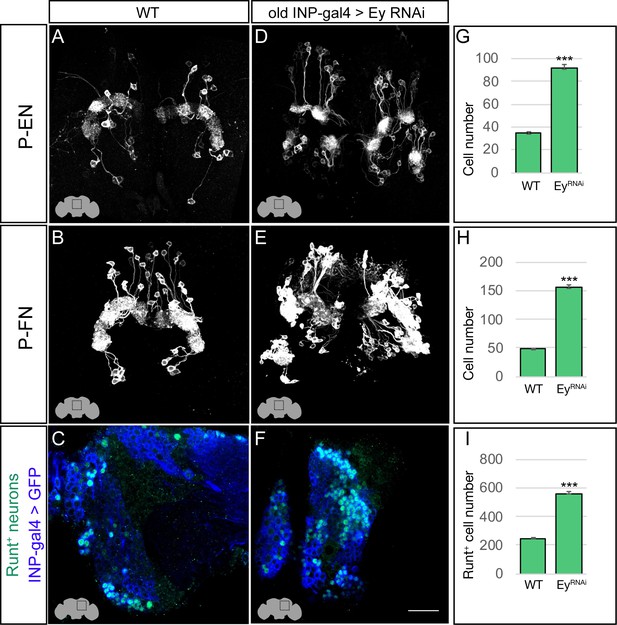
Eyeless represses P-EN and P-FN molecular identity.
(A–C) Wild-type P-EN, P-FN, and Runt+ neurons in the dorsoposterior adult brain. P-EN and P-FN neurons detected by expression of neuron-specific LexA lines. See Materials and methods for genotypes. (D–F) EyelessRNAi in INP lineages increases the number of P-EN, P-FN, and Runt+ late-born neurons in the dorsoposterior adult brain. See Materials and methods for genotypes. (G–I) Quantification (n = 5 for each experiment). ***, p<0.001. Scale bar, 20µm.
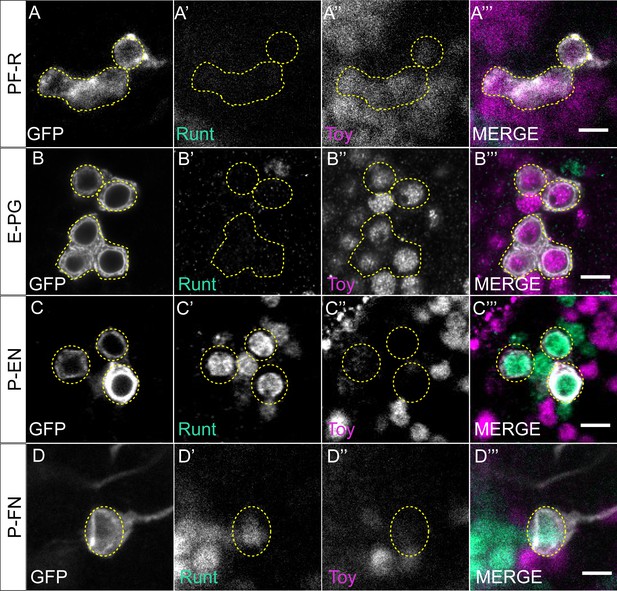
CX columnar neurons express either Runt or Toy in adult brain.
(A–A’’’) Cell bodies of PF-R neurons in the adult brain express the TF Toy but not Runt, as labeled by R37G12-lexA. Scale bar, 5 μm. (B–B’’’) Cell bodies of E-PG neurons in the adult brain express the TF Toy but not Runt, as labeled by R60D05-lexA. Scale bar, 5 μm. (C–C’’’) Cell bodies of P-EN neurons in the adult brain express the TF Runt but not Toy, as labeled by R12D09-lexA. Scale bar, 5 μm. (D–D’’’) Cell bodies of P-FN neurons in the adult brain express the TF Runt but not Toy, as labeled by R16D01-lexA. Scale bar, 5 μm.
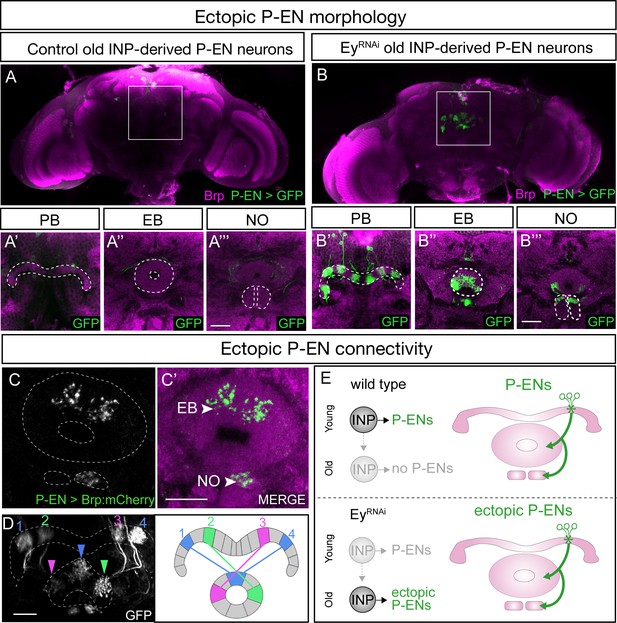
EyelessRNAi produces late-born ‘ectopic’ P-EN neurons that have normal P-EN morphology and connectivity.
(A–A’’’) In wild-type adults, late INP clones do not label P-EN neurons in the adult brain (n = 5). See Materials and methods for details. PB, EB, and NO neuropils marked with dashed lines. Scale bars, 20µm (A–B). (B–B’’’) In EyelessRNAi adults, late INP clones produce ectopic late-born P-EN neurons, which project to the PB, EB, and Noduli (n = 5), similar to endogenous P-EN neurons (Figure 1A,B). PB, EB, and NO neuropils marked with dashed lines. (C–C’) In EyelessRNAi adults, late INP clones produce ectopic late-born P-EN neurons, which localize the pre-synaptic marker Brp::mCherry to the EB and Noduli (n = 5), but not to the PB (not shown), similar to the endogenous P-EN neurons. Scale bars, 20µm (C–D). (D) EyelessRNAi adult, showing stochastic labeling of four ectopic P-EN neurons (1-4) with normal PB and EB glomeruli targeting (compare to Wolff and Rubin, 2018). (E) Summary.
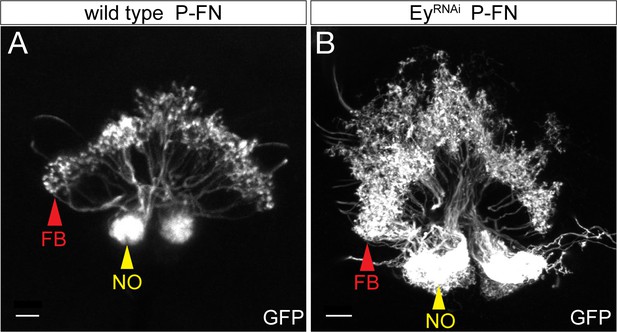
EyelessRNAi produces late-born ‘ectopic’ P-FN neurons that have normal P-FN morphology.
(A) WT P-FN neurons innervate the FB (red arrowhead) and the NO (yellow arrowhead), as labeled by R16D01-lexA. (B) EyelessRNAi generates ectopic P-FN neurons, resulting in increased innervation of the FB and NO (red and yellow arrowheads respectively), creating enlarged neuropil structures. Indicating that ectopic copies of P-FN neurons generated by EyelessRNAi have the same morphological targeting as endogenous copies of P-FN neurons.
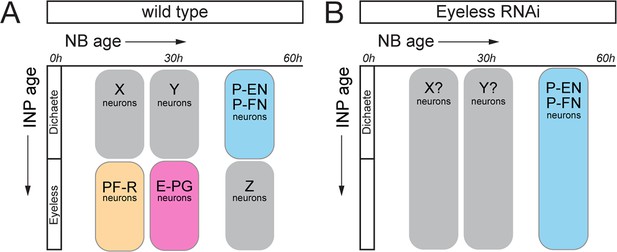
Eyeless regulation of identity schematic.
(A) Birth-dating results demonstrate that CX columnar neurons are generated by discrete temporal windows within DM1-DM4 type II neuroblast lineages. (B) EyelessRNAi results in the elimination of old INP born neurons such as PF-R and E-PG neurons, and subsequent expansion of young INP neurons such as P-EN and P-FN neurons.
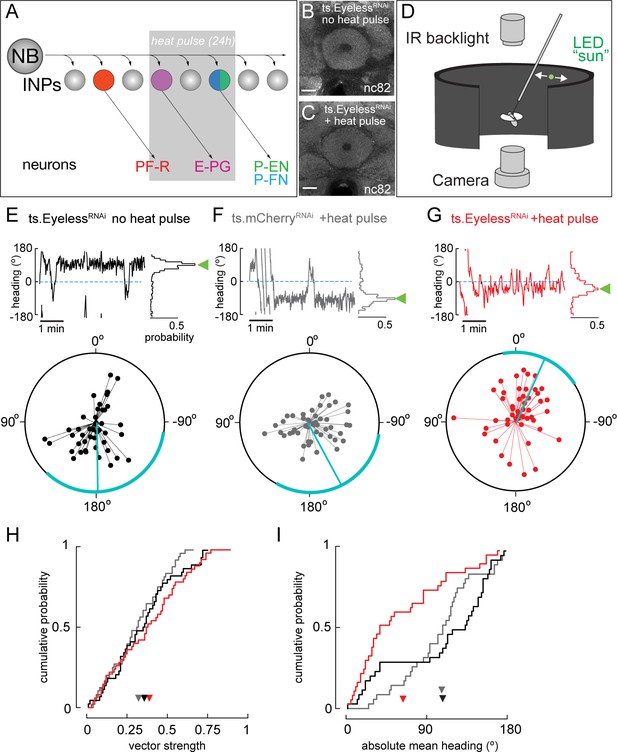
Transient loss of Eyeless during development impairs adult fly navigation.
(A) Timing of Eyeless reduction in INP lineages. Transient inactivation of ts.Gal80 (29°C heat pulse; gray bar) results in transient EyelessRNAi during the time in which E-PG neurons are normally generated. (B,C) The manipulation in A does not alter CX neuropil morphology as seen by nc82 (neuropil) staining (EB, shown; other neuropils, data not shown). (D) Schematic of experimental apparatus for sun navigation experiments. The wing stroke amplitudes of a tethered, flying fly were monitored with an IR camera; the stroke difference determined the angular velocity of a 2.4° sun stimulus. Modified from (Giraldo et al., 2018). (E) Example flight (top panel) and summary data (bottom panel) from ts.EyelessRNAi control with no heat pulse. Top panel: left plot shows headings over 5 min flight; 0° is sun position in front of fly. Right histogram is distribution of headings in this example flight; sideways green triangle is the mean. Bottom panel: summary data, with each 5 min flight represented by radial lines. The angle of each line is the mean flight heading. The length of each line is vector strength of flight, varying from 0 (center of circle; no stimulus stabilization) to 1 (edge of circle; perfect stabilization). Each fly flew for two 5 min flights separated by a 5 min rest period. Cyan line shows mean heading, across flights with vector strength >0.2, as well as 95% confidence interval, calculated via resampling across flies. 44 flights, 22 flies total. (F) Example and summary data from ts.mcherryRNAi control. Same plotting convention as (E). 48 flights, 24 flies. (G) Example and summary data from ts.EyelessRNAi flies with heat pulse. Same plotting convention as (E,F). 50 flights, 25 flies. (H) Cumulative probability distribution of vector strengths from both control groups (black and gray) and from experimental group (red). There was no significant difference between means (ts.EyelessRNAi heat pulse, 0.34, red; ts.EyelessRNAi no heat pulse, 0.31,black;ts.mcherryRNAi heat pulse 0.30,gray; p>0.1, permutation test). (I) Cumulative probability distribution of mean absolute headings. The heading distribution for the experimental group, ts.EyelessRNAi heat pulse, was skewed significantly to frontal headings (mean 63.7°, 37 flights in 23 flies with vector strength >0.2) compared to control distributions (p<0.01, permutation test; ts.EyelessRNAi no heat pulse, mean 107.9°, 35 flights in 19 flies; ts.mcherryRNAi, mean 106.9°, 31 flights in 21 flies). Scale bars, 10µm.
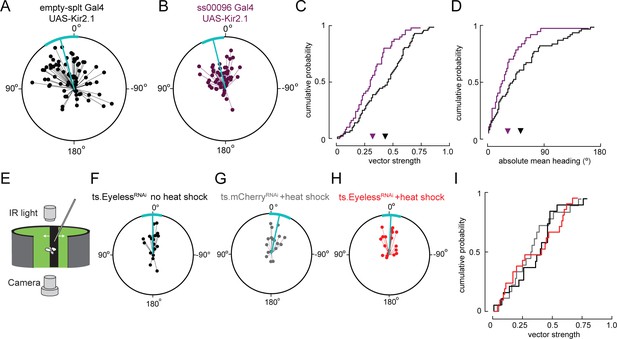
Sun navigation is impaired when E-PG neurons are silenced; stripe fixation behavior is not altered by the loss of Eyeless.
(A–D) Silencing E-PG neurons impairs the flies’ capacity to maintain an arbitrary heading to a bright spot resembling the sun. (A) Flight headings to sun stimulus in empty-Split/UAS Kir 2.1 control flies. Plotting convention same as Figure 7E–G. N = 72 flights, 36 flies. (B) Flight headings to sun stimulus in ss00096 Split-Gal4 (E-PG neurons)/UAS-Kir2.1 flies. N = 50 flights, 25 flies. (C) Cumulative probability distribution of vector strength values for flights to sun stimulus. empty Split-Gal4 UAS-Kir2.1 (black line), mean = 0.43, N = 72 flights, 36 flies; ss00096 Split-Gal4/UAS-Kir2.1 (purple line), mean = 0.32, N = 50 flights, 25 flies. p=0.001, permutation test for significant difference in mean. (D) Cumulative probability distribution of mean absolute heading for flights to sun stimulus. empty Split-Gal4 UAS-Kir2.1 (black line), mean = 52.7°, N = 61 flights, 36 flies; ss00096 Split-Gal4/UAS-Kir2.1 (purple line), mean = 32.2°, N = 37 flights, 23 flies. p=0.011, permutation test for significant difference in mean. (E–H) Transient removal of Eyeless in larvae at the time of E-PG development (as in Figure 7) has no effect on stripe fixation behavior in adult flies. Plotting conventions as in Figure 7E–G. (E) Schematic of behavioral apparatus; flies controlled rotation of a dark vertical stripe with their wing strokes. (F) Control ts.EyelessRNAi no heat pulse. N = 19 flies. Flight headings are tightly clustered around 0°, where stripe is in front of fly. (G) Control ts.mcherryRNAi flies with heat pulse. N = 18 flies. (H) Experimental ts.EyelessRNAi flies with heat pulse. Heading distribution remains centered around 0°. N = 21 flies. (I) Cumulative probability distribution of vector strength values for stripe stimuli with control groups (black and gray) and experimental group (red). We observed no significant difference between means for all genotypes.
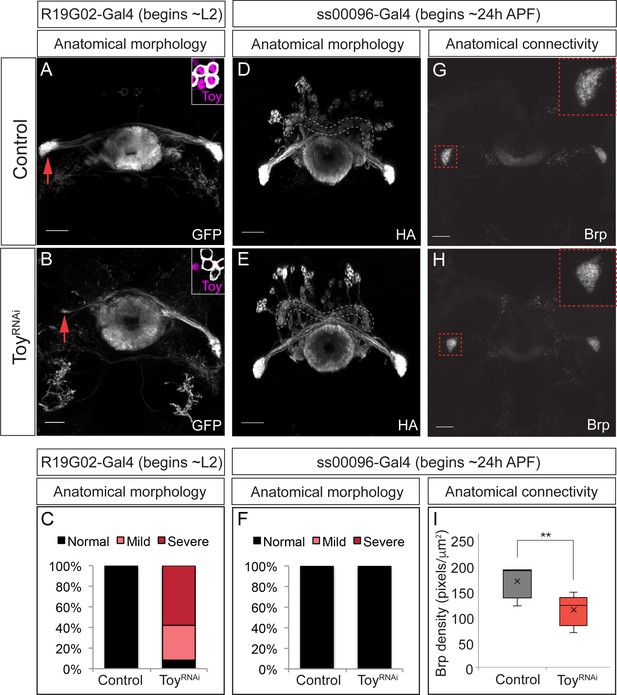
The Eyeless target gene Toy is required for E-PG axonal connectivity.
(A–C) Loss of Toy in larvae reduces E-PG projections to the Gall in adults. (A) Wild type: R19G02-Gal4 is first expressed at ~L2 and labels adult E-PG neurons; note projections to the EB (center) and Gall (left and right); PB, not shown (n = 6). Inset shows WT levels of Toy-protein expression. (B) R19G02-Gal4 UAS-ToyRNAi reduces E-PG projections to the Gall (red arrow), yet projections to the EB and PB (not shown) remain intact (n = 12). Inset shows loss of Toy-protein expression. Quantification: mild, detectable reduction in the Gall in one hemisphere; severe, virtually complete loss of Gall. (D–F) Loss of Toy in pupae has no effect on E-PG projections. (D) In wild type, ss00096 split-Gal4 is expressed ~24 hr after pupal formation and labels adult E-PG neurons; n = 5. (E) ss00096 split-Gal4 UAS-ToyRNAi adults have normal projections to the EB (center), Gall (left and right), and PB (outlined); n = 5. (F) Quantification. (G–I) Loss of Toy in pupae reduces pre-synaptic levels of Brp in the Gall. (G,H) Genotypes as in D-E, showing that the pre-synaptic marker Brp is reduced in the E-PG axons targeting the Gall following ToyRNAi (n = 6). (I) Quantification. Scale bars, 20µm.
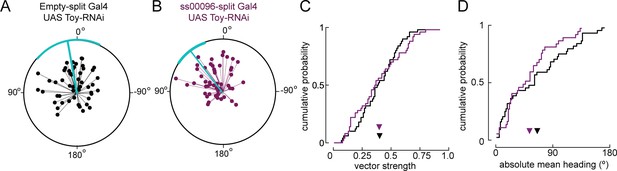
Reducing levels of Toy in E-PG neurons during pupal stages does not alter navigation to a fictive sun.
(A) Flight headings to sun stimulus in empty-split Gal4/UAS Toy-RNAi control flies. As in Figure 7—figure supplement 1, each 5 min flight is represented with a distinct radial line. The angle of the line is the mean flight heading and the length of line is vector strength. The cyan line shows mean heading across flights with vector strength >0.2 as well as a 95% confidence interval, calculated via resampling across flies. N = 72 flights, 36 flies. (B) Flight headings to sun stimulus in ss00096-split Gal4 (E-PG neurons)/UAS Toy-RNAi flies. Plotting convention as in (A). N = 42 flights, 21 flies. (C) Comparison of cumulative probability distribution of vector strength values. Control empty-split Gal4 UAS Toy-RNAi (black line), mean = 0.40; ss00096-split Gal4/UAS Toy-RNAi (purple line), mean = 0.40. (D) Cumulative probability distribution of mean absolute heading for flights to sun stimulus. empty-split Gal4 UAS/Toy RNAi (black line), mean = 65.4°, N = 44 flights, 25 flies; pupal ss00096-split Gal4/UAS Toy-RNAi (purple line), mean = 53.2°, N = 37 flights, 21 flies. Permutation test for significant difference of means, p=0.17.
Videos
Wild-type adult P-EN morphology wild-type young INP born P-EN neurons innervate distinct neuropil regions of the central complex.
These include the PB, EB, and NO outlined in red, white, and yellow respectively.
Ey-RNAi ectopic adult P-EN morphology Ey-RNAi ectopic old INP born P-EN neurons innervate the same distinct neuropil regions of the central complex.
These include the PB, EB, and NO outlined in red, white, and yellow respectively. Note, only even-numbered glomeruli are generated, indicating they are born after odd numbered glomeruli in the INP lineage, and extend when Eyeless is eliminated in old INPs.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Species (Drosophila melanogaster) | UAS-FLP | BDSC | #4539 | FLP enzyme under UAS control |
| Species (Drosophila melanogaster) | R9D11-Gal4 | BDSC | #40731 | Young INP Gal4 driver |
| Species (Drosophila melanogaster) | R37G12-lexA | BDSC | #52765 | PF-R lexA driver |
| Species (Drosophila melanogaster) | R60D05-lexA | BDSC | #52867 | E-PG lexA driver |
| Species (Drosophila melanogaster) | R12D09-lexA | BDSC | #54419 | P-EN lexA driver |
| Species (Drosophila melanogaster) | R16D01-lexA | BDSC | #52503 | P-FN lexA driver |
| Species (Drosophila melanogaster) | lexAop(FRT.stop)mCD8::GFP | BDSC | #57588 | FLP-out membrane bound GFP under lexAop control |
| Species (Drosophila melanogaster) | ts.Tubulin-Gal80 (20) | BDSC | #7019 | temperature sensitive Gal80 |
| Species (Drosophila melanogaster) | 20xUAS-FLP.PEST | BDSC | #55807 | FLP enzyme under 20xUAS control |
| Species (Drosophila melanogaster) | OK107-Gal4 | BDSC | #854 | Eyeless enhancer trap Gal4 for old INPs |
| Species (Drosophila melanogaster) | UAS-mCherryRNAi | BDSC | #35787 | Control RNAi under UAS control |
| Species (Drosophila melanogaster) | UAS-EyelessRNAi | BDSC | #32486 | Eyeless RNAi under UAS control |
| Species (Drosophila melanogaster) | 13xlexAop-myr::GFP | BDSC | #32210 | membrane bound GFP under 13xlexAop control |
| Species (Drosophila melanogaster) | R16B06-Gal4 | BDSC | #45811 | old INP Gal4 driver |
| Species (Drosophila melanogaster) | UAS-FLP, Act5c(FRT.CD2)Gal4; ; R12E09-Gal4, UAS-mCD8::GFP | This work | INP immortalization driver expressing membrane bound GFP | |
| Species (Drosophila melanogaster) | lexAop(FRT.stop)HA::CD4.T2A.Brp.mCherry | BDSC | #56518 | FLP-out fluorensent labeling of Brp |
| Species (Drosophila melanogaster) | ss00096-Gal4 | Rubin Lab (Janelia) | E-PG split Gal4 driver | |
| Species (Drosophila melanogaster) | Empty-vector split Gal4 | Rubin Lab (Janelia) | Control split Gal4 driver | |
| Species (Drosophila melanogaster) | UAS-ToyRNAi | BDSC | #33679 | Toy RNAi under UAS control |
| Species (Drosophila melanogaster) | lexAop.tdTomato.myr, brp(FRT.stop)V5-2A-lexA-VP16 | BDSC | #56142 | STaR FLP-out labeling of synaptic terminals |
| Species (Drosophila melanogaster) | 10xUAS-myr::HA | BDSC | #62145 | membrane bound HA under UAS control |
| Species (Drosophila melanogaster) | R19G02-Gal4 | BDSC | #48860 | developmental E-PG Gal4 |
| Species (Drosophila melanogaster) | UAS-Kir2.1 | Giraldo et al., 2018 | Inward rectifying K + channel under UAS control | |
| Antibody, polyclonal | Chicken anti-GFP | Abcam (Eugene, OR) | 1:1000 | |
| Antibody, polyclonal | Rabbit anti-Toy | Desplan lab (NYU) | 1:1000 | |
| Antibody, polyclonal | Guinea-pig anti-Runt | Desplan lab (NYU) | 1:1000 | |
| Antibody, monoclonal | Mouse anti-nc82 | DSHB (Iowa City, IA) | 1:50 | |
| Antibody, polyclonal | Rabbit anti-V5 | Cell Signaling (Danvers MA) | 1:400 | |
| Antibody, polyclonal | Rabbit Anti HA | Columbia Biosciences (Frederick MD) | 1:400 | |
| Antibody, polyclonal | Secondary antibodies | Thermofisher (Eugene, OR) | 1:400 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43482.019





