Human macular Müller cells rely more on serine biosynthesis to combat oxidative stress than those from the periphery
Figures
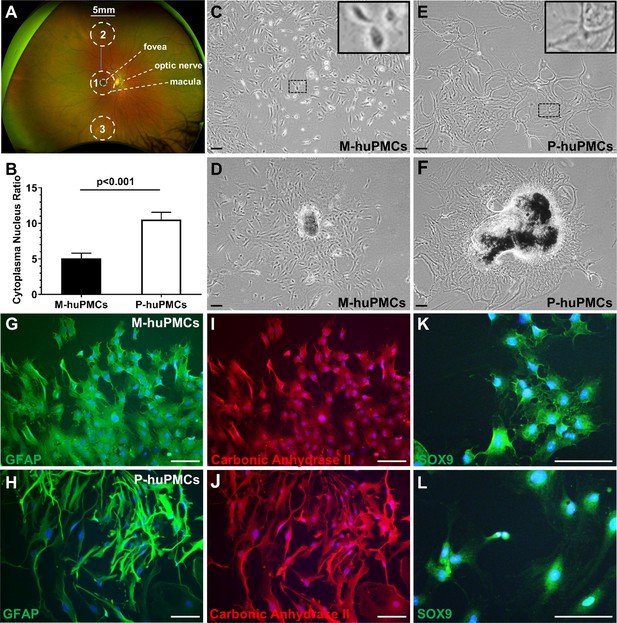
Morphology of primary macular and peripheral Müller cells.
(A) A wide-field fundus image showing the retinal areas used for primary Müller cell culture. Retinal tissue at Area one was used to derive primary Müller cells from the macula while the punches from Areas 2 and 3 were pooled to derive primary Müller cells from the peripheral retina. (B) Cytoplasm/nucleus ratio of primary Müller cells from the macula (M-huPMCs) and peripheral (P-huPMCs) human retina (n = 8). (C-F) Bright field images of huPMCs isolated from the macula (C-D) or peripheral (E-F) retina. (D) Primary Müller cells that grew out from macular retinal piece. (F) Primary Müller cells that grew out from peripheral retinal piece. (G-L) M-huPMCs (upper panel) and P-huPMCs (lower panel) (P0) without sub-culturing expressed three specific protein markers of Müller cells: GFAP (G-H), carbonic anhydrase II (I-J) and SOX9 (K-L). Black scale bar = 200 µM, white scale bar = 100 µM.
-
Figure 1—source data 1
Source data for Figure 1B.
- https://doi.org/10.7554/eLife.43598.004
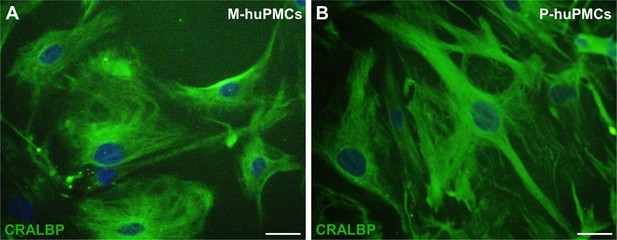
CRALBP immunostaining of M-huPMCs and P-huPMCs.
M-huPMCs (A) and P-huPMCs (B) at P0 passage both expressed CRALBP, the specific protein marker of Müller cells. Scale bar = 50 µM (M = macula; p=peripheral).
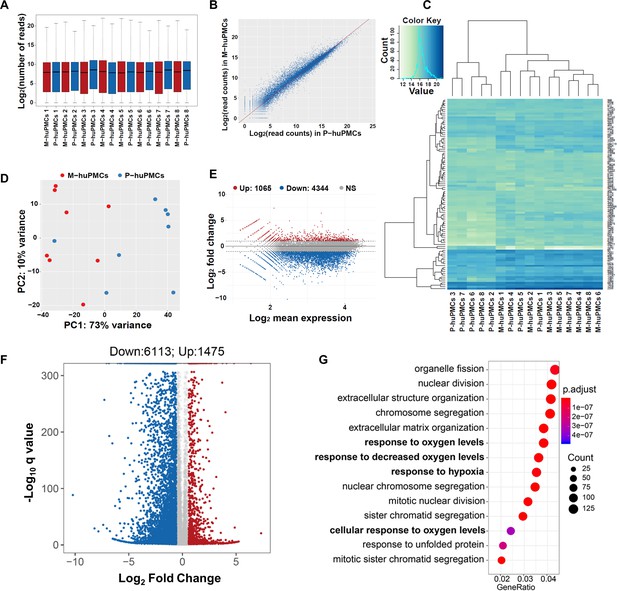
Transcription profiles of macular and peripheral Müller cells.
(A) Boxplot of the Reads expression values of the M-huPMCs and P-huPMCs cultured from eight donor retinas. (B) Scatter Plot for correlations of genes expressed in the M-huPMCs and P-huPMCs. (C) Heatmap of differentially expressed genes between the M-huPMCs and P-huPMCs of eight donors shows the top 100 genes with the smallest q-values. (D) Principal component analysis (PCA) was performed with the RNA-seq data derived from the M-huPMCs and P-huPMCs. (E) Bland–Altman (MA) plot of differentially expressed genes in the M-huPMCs and P-huPMCs. (F) Volcano plot of differentially expressed genes in the M-huPMCs and P-huPMCs. The log 10 (q values) were plotted against the log 2 (Fold Change) in gene expression. The genes upregulated (n = 1475) 1.5-fold or more and with an FDR corrected p-value<0.05 were depicted as red dots; while the genes downregulated (n = 6113) by 1.5-folds or more and with a FDR corrected p-value<0.05 were depicted as blue dots. (G) Gene ontology (GO) analysis of upregulated genes in M-huPMCs relative to P-huPMCs. The top 14 GO terms in the biological process category are displayed and ordered by enriched gene number and adjusted p value.
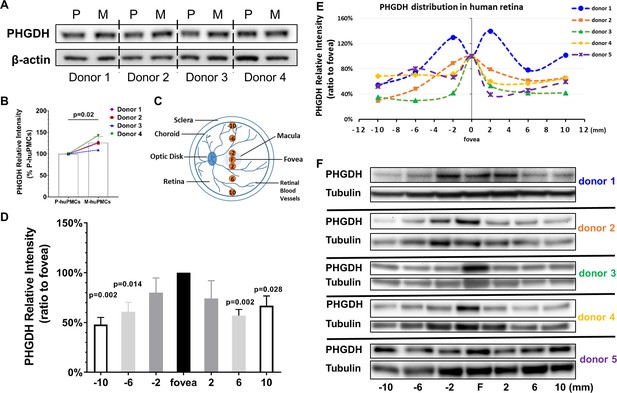
PHGDH expression in cultured Müller cells and retinal tissue from macula and periphery.
(A) Representative immunoblot showing PHGDH protein expression in primary cultures of human Müller cells from macula and peripheral retina; (B) Quantitative analysis of PHGDH relative to β-actin for immunoblots (n = 4 donor retinas); (C) Schema of trephined retina area used for Western blotting shown in (F); (D-E) Quantitative analysis of the expression of PHGDH correlated to different areas of human retina (five donors); (F) Expression of PHGDH in different retinal locations shown using western blotting (n = 5 donors).
-
Figure 3—source data 1
Source data for Figure 3B,D,E,F.
- https://doi.org/10.7554/eLife.43598.010
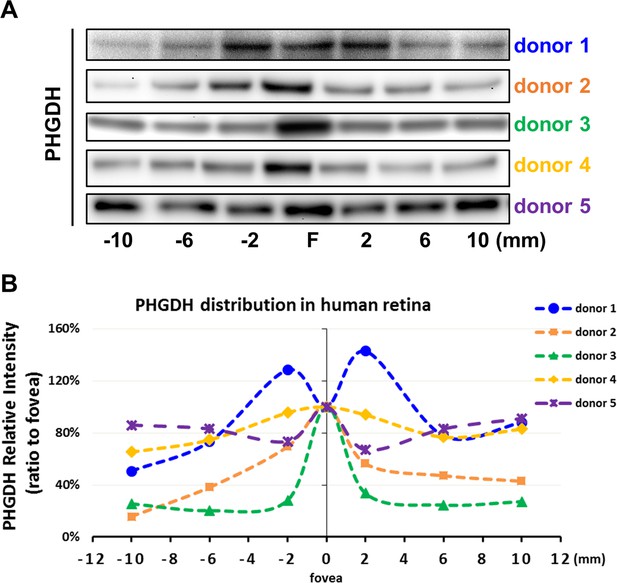
PHGDH expression in the macula and peripheral retina.
(A) PHGDH expression in different retinal locations was assessed with western blot (n = 5 donors). (B) Quantitative analysis of PHGDH expression (normalized to total loaded retinal protein) was correlated with different areas of human retina (n = 5 donors).
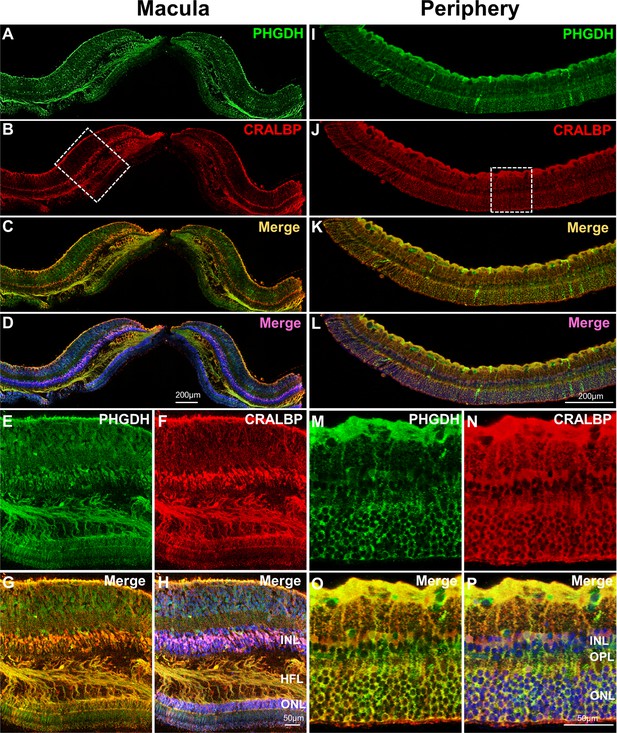
Immunofluorescent staining of PHGDH in human retina.
Representative images of immunofluorescence visualization of PHGDH (green) and CRALBP (red, a Müller cell marker) in human macular (A-H) and peripheral retina (I-P). E-H Field enlarged image from B (white-dotted box) in macula; M-P. Enlarged images from J (white-dotted box) in peripheral retina. The specific immunoreactivity of PHGDH antibody was verified in PHGDH knockdown in MIO-M1 cells (Figure 4—figure supplement 1).
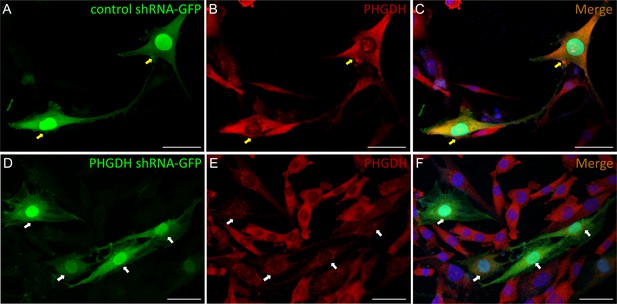
Verification of PHGDH specific immunoreactivity by PHGDH knockdown in MIO-M1 cells.
Cells expressing green fluorescence have been successfully transfected with control (A) or PHGDH (D) shRNA-GFP plasmid. PHGDH was immunostained with anti-PHGDH antibody, visualised with Alexa 594 (red) in the MIO-M1 cells (B and E). (C) Merged images of A and B showing normal PHGDH immunoreactivity in cells transfected with negative control shRNA-GFP plasmid (yellow arrows). (F) Merged images of D and E showing reduced PHGDH immunoactivity in the cells transfected with PHGDH-shRNA-GFP plasmid (white arrows). Scale bar = 50 µM.
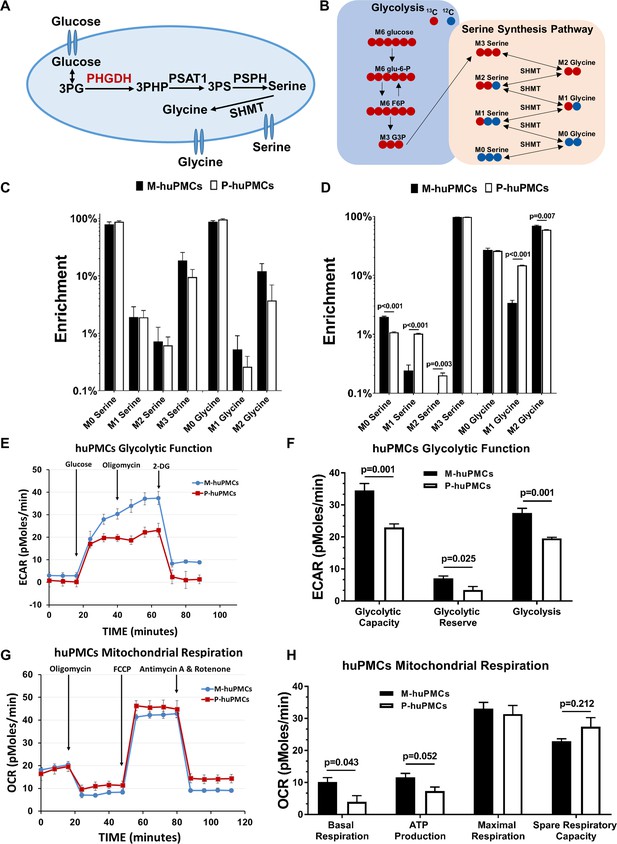
Metabolic differences between the macular and peripheral Müller cells.
(A) de novo serine/glycine synthesis pathway. 3PG: 3-phosphoglycerate, PHGDH: 3-phosphoglycerate dehydrogenase, PSAT1: phosphoserine aminotransferase 1, PSPH: phosphoserine phosphatase, SHMT: serine hydroxymethyltransferase. (B) Simplified schematic of steps in de novo serine/glycine synthesis, showing 13C labeling patterns resulting from 13C6 glucose substrate. Red fills indicate 13C atoms. (C–D) 13C-serine/glycine levels in M-huPMCs and P-huPMCs after treatment with 13C-glucose (C) or 13C-serine (D). (E–H) Seahorse XF analysis of M-huPMCs and P-huPMCs. Glycolytic (E–F) and mitochondrial functions (G–H) were evaluated in human Müller cells in primary culture isolated from macula and peripheral regions (n = 4).
-
Figure 5—source data 1
Source data for Figure 5C,D,F,H.
- https://doi.org/10.7554/eLife.43598.015
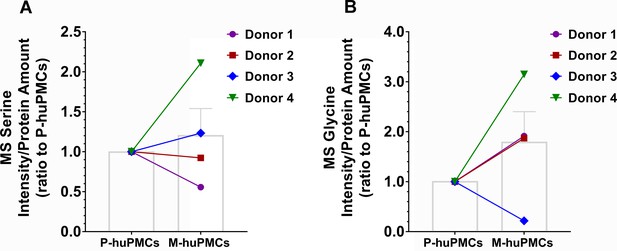
The ratio of serine (A) or glycine (B) MS intensity to total amount of cellular protein in M-huPMCs and P-huPMCs from the same donor.
https://doi.org/10.7554/eLife.43598.014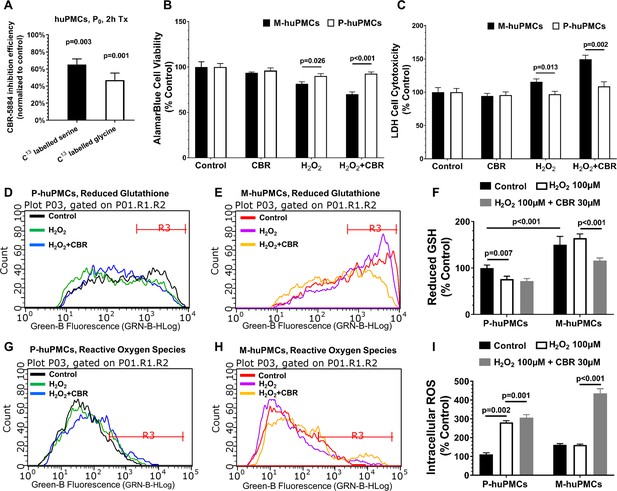
Responses to oxidative stress of primary cultured Müller cells from macula and peripheral retina.
(A) Measurement of 13C-serine and -glycine after PHGDH inhibition in human primary Müller cells; B-C. Cell metabolic activity (B) and cellular damage (C) in the M-huPMCs and P-huPMCs with or without PHGDH inhibition under oxidative stress; D-I. Flow cytometry analysis for M-huPMCs and P-huPMCs stained with Thiol Green Dye to detect GSH (D-F) and with CM-H2DCFDA to evaluate ROS (G-I) for cells under mild oxidative stress (100 μM H2O2), with or without PHGDH inhibition (n = 5).
-
Figure 6—source data 1
Source data for Figure 6A–C,F,I.
- https://doi.org/10.7554/eLife.43598.018
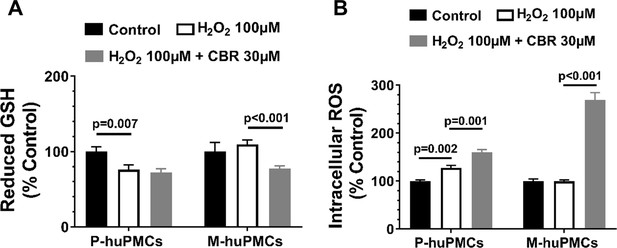
Quantitative re-analysis of the flow cytometry data in Figure 6D–E,G–H.
(A-B)ROS (CM-H2DCFDA) and GSH (Thiol Green Dye) levels in M-huPMCs and P-huPMCs in response to induced hyperoxia with and without CBR, an inhibitor of PHDGH, relative to untreated controls (n = 5 per group).
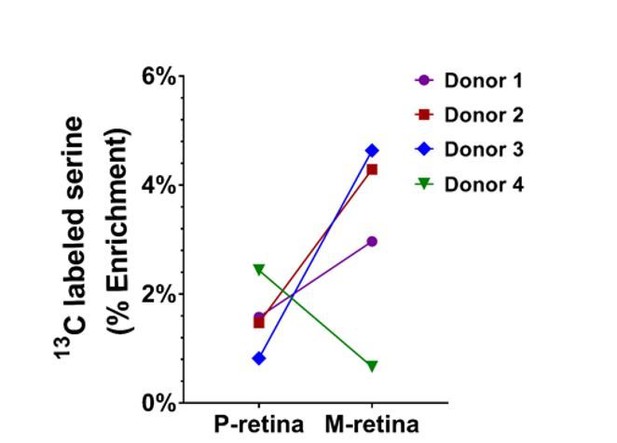
13C-labelled serine level in the macula and peripheral retina following the treatment of 13C-glucose (n=4 donors).
Donors 1, 2 and 3 showed a higher 13C-labelled serine level in the macula vs peripheral retina. Donor 4 showed reduced 13C-labelled serine in the macula.
Tables
Gene expression of retinal cells marker in identified cells from macula and peripheral retina
https://doi.org/10.7554/eLife.43598.006| Marker | Gene name | Gene ID | Length | M-rpkm | P-rpkm | Log2ratio(P/M) | Regulation(P/M) | p-value |
|---|---|---|---|---|---|---|---|---|
| Müller cell | Vimentin | 7431 | 2151 | 2923.82 | 3492.22 | 0.26 | up | <2.22E-308 |
| Glutamine synthetase | 2752 | 4737 | 23.46 | 95.31 | 2.02 | up | <2.22E-308 | |
| RLBP1 (CRALBP) | 6017 | 1752 | 4.50 | 34.79 | 2.95 | up | <2.22E-308 | |
| CLU clusterin | 1191 | 3012 | 78.80 | 763.72 | 3.28 | up | <2.22E-308 | |
| Carbonic anhydrase II | 760 | 1666 | 15.52 | 138.45 | 3.16 | up | <2.22E-308 | |
| GFAP | 2670 | 3097 | 31.87 | 944.58 | 4.89 | up | <2.22E-308 | |
| Other cell types | ||||||||
| Photoreceptors | RHO rhodopsin | 6010 | 2768 | 0.01 | 0.05 | 2.13 | up | 1.43E-07 |
| Biopolar cells | mGluR6 | 2916 | 6143 | 0.03 | 0.31 | 3.2 | up | 3.13E-134 |
| Amacrine cells | STX1A syntaxin 1A | 6804 | 2138 | 4.35 | 3.89 | −0.16 | down | 3.43E-06 |
| Microglia | Iba1 | 199 | 1491 | 1.44 | 3.67 | 1.35 | up | 3.99E-129 |
| Endothelial | PECAM1 | 5175 | 6831 | 0.25 | 0.72 | 1.52 | up | 1.21E-138 |
Serine synthesis pathway-related gene expression in Müller cell isolated from macula and peripheral retina.
https://doi.org/10.7554/eLife.43598.007| Gene name | Gene ID | Length | M-rpkm | P-rpkm | Log2ratio (M/P) | Regulation (M/P) | p-value |
|---|---|---|---|---|---|---|---|
| PHGDH | 26227 | 2021 | 60.36 | 40.09 | 0.59 | up | <2.22E-308 |
| PSAT1 | 29968 | 2221 | 101.15 | 79.10 | 0.35 | up | <2.22E-308 |
| PSPH | 5723 | 2142 | 12.51 | 14.20 | −0.18 | down | 2.56E-21 |
| SHMT1 | 6470 | 2540 | 3.72 | 7.05 | −0.92 | down | 2.99E-228 |
| SHMT2 | 6472 | 2357 | 64.83 | 61.14 | 0.08 | up | 1.27E-23 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit polyclonal anti-3-PGDH | Millipore | Cat# ABS571, RRID:AB_2783876 | IF(1:1000), WB (1:1000) |
| Antibody | Rabbit polyclonalanti-α/β tubulin | Cell Signaling Technology | Cat# 2148, RRID:AB_2288042 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-CRALBP | Abcam | Cat# ab15051, RRID:AB_2269474 | IF(1:500) |
| Antibody | Goat polyclonalanti-GFAP | Abcam | Cat# ab53554, RRID:AB_880202 | IF(1:1000) |
| Antibody | Rabbit polyclonal anti-carbonicanhydrase II | Abcam | Cat# ab6621, RRID:AB_305602 | IF(1:1000) |
| Antibody | Rabbit polyclonal anti-Sox9 | Millipore | Cat# AB5535, RRID:AB_2239761 | IF(1:1000) |
| Recombinant DNA reagent | PHGDH-shRNA-GFP plasmid | Sigma Aldrich | MISSION SHRNA PLASMID DNA NM_006623/TRCN0000233032 / -hPGK-Neo - CMV-tGFP | |
| Recombinant DNA reagent | Negative control-shRNA-GFP plasmid | Sigma Aldrich | MISSION SHRNA CUSTOM DNA NEGATIVE CONTROL - nontarget shRNA (SHC016) - pLKO.1-Neo-CMV-tGFP vector | |
| Commercial assay or kit | Intracellular GSH detection kit | Abcam | ab112132 | |
| Commercial assay or kit | CM-H2DCFDA | Molecular Probes | C6827 | |
| Commercialassay or kit | Pierce LDH cytotoxicity assay kit | ThermoFisher Scientific | 88953 | |
| Commercialassay or kit | AamarBlue cellviability reagent | ThermoFisher Scientific | DAL1100 | |
| Chemicalcompound, drug | CBR-5884 | Sigma Aldrich | SML1656 | |
| Software,algorithm | SPSS | SPSS | RRID:SCR_002865 |
Additional files
-
Supplementary file 1
RNA sequencing raw data.
- https://doi.org/10.7554/eLife.43598.019
-
Supplementary file 2
RNA sequencing filtered data.
- https://doi.org/10.7554/eLife.43598.020





