A novel class of inferior colliculus principal neurons labeled in vasoactive intestinal peptide-Cre mice
Figures
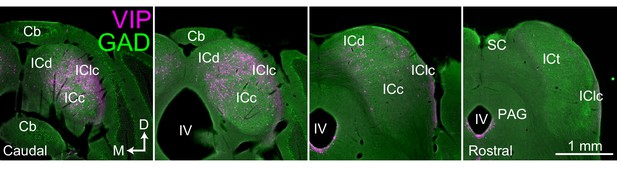
VIP neurons are distributed across multiple subdivisions of the IC.
Photomicrographs of transverse sections through the IC ranging from caudal (left-most) to rostral (right-most). VIP-expressing cells (labeled with tdTomato) are shown in magenta, and GAD67 staining is shown in green to show the border of the IC. VIP-expressing cells are present in multiple subdivisions of the IC, but are most prominent in caudal and dorsal parts of the IC. Scale = 1 mm. Cb (cerebellum), ICc, ICd, IClc (central nucleus, dorsal cortex and lateral cortex of the inferior colliculus), ICt (intercollicular tegmentum), IV (fourth ventricle), PAG (periaqueductal gray).
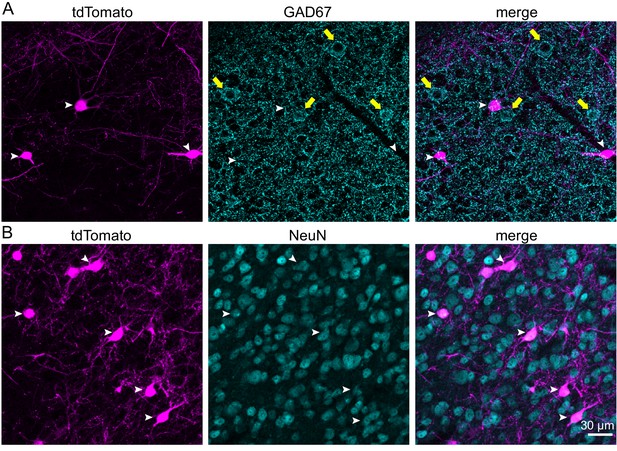
VIP neurons are glutamatergic and represent 3.5% of neurons in the ICc.
(A) Confocal z-stack projections showing IC VIP neurons (magenta, left), GAD67 staining (cyan, middle), and an overlay (right). White arrowheads mark VIP neurons, yellow arrows GABAergic cell bodies. There was virtually no overlap between VIP neurons and GABAergic neurons (right). (B) Confocal z-stack projections showing VIP neurons (magenta, left), NeuN staining (cyan, middle), and an overlay (right). White arrowheads mark VIP neurons labeled by NeuN. Scale bar applies to A and B.
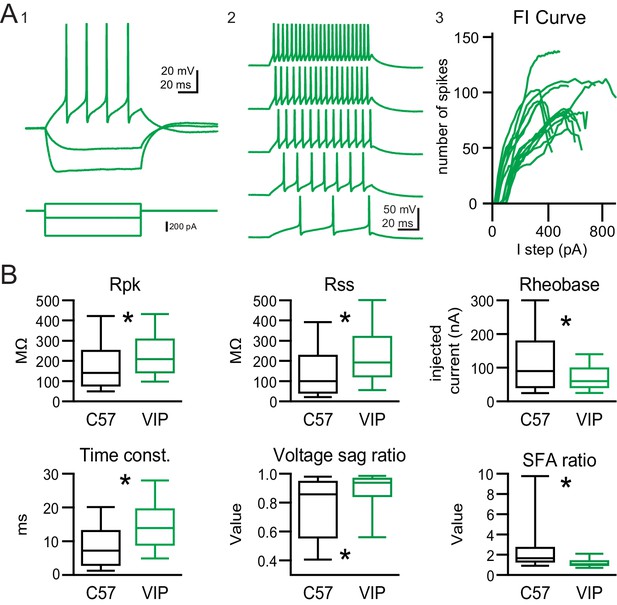
VIP neurons have sustained firing patterns and moderate membrane properties.
(A) VIP neurons exhibited a regular, sustained firing pattern in response to depolarizing current steps, while hyperpolarizing current steps elicited minimal voltage sag (A1). As the amplitude of depolarizing current steps was increased, VIP neurons increased their firing rate while keeping their sustained firing pattern (A2). Example firing versus input (FI) curves from 15 VIP neurons show that firing rate increased in a mostly linear fashion over a broad range of current step amplitudes (A3). (B) Intrinsic physiology of VIP neurons is statistically different from the general population of IC neurons for all parameters tested. On average, VIP neurons had a significantly higher peak input resistance (Rpk) and steady-state input resistance (Rss), a lower rheobase, a longer membrane time constant, a smaller and less variable voltage sag (Ih) at −91 mV, and a markedly small and highly invariable spike frequency adaptation ratio (SFA). Boxplots show median, 25th and 75th percentile (box), and 9th and 91th percentile (whiskers).
-
Figure 3—source data 1
Intrinsic physiology of VIP neurons and from non-targeted recordings in mouse IC.
- https://doi.org/10.7554/eLife.43770.009
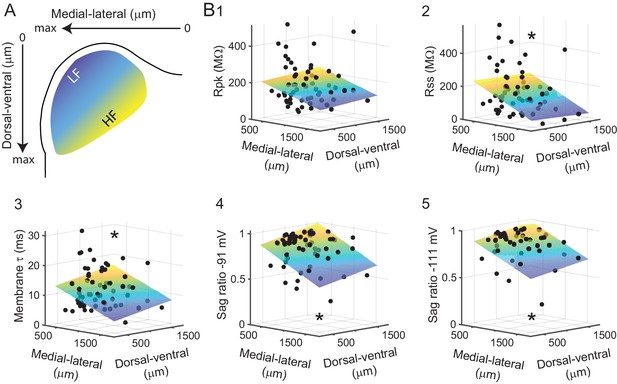
Intrinsic physiology of VIP neurons in the ICc varies along the tonotopic axis.
(A) A 2D coordinate system was fit to every IC slice a VIP neuron was recorded from. The medial-lateral axis runs from the midline (zero) to the lateral edge (max) of the slice, the dorsal-ventral axis from the dorsal edge of the slice (zero) to the ventral border of the IC (max). For illustrative purposes, the approximate position along the tonotopic axis of the ICc is color-coded from blue (low frequency) to yellow (high frequency). (B) Correlation of measured intrinsic parameters with recording location. Black dots represent physiological parameters of individual VIP neurons (z-axis, left) mapped to their recording location (x- and y-axes, bottom). Planes show Levenberg-Marquardt least squares fits, color-coded from low z-axis values (blue) to high z-axis values (yellow). Asterisks indicate statistical significance of fit.
-
Figure 4—source data 1
Intrinsic physiology of VIP neurons matched to their location in the ICc.
- https://doi.org/10.7554/eLife.43770.011
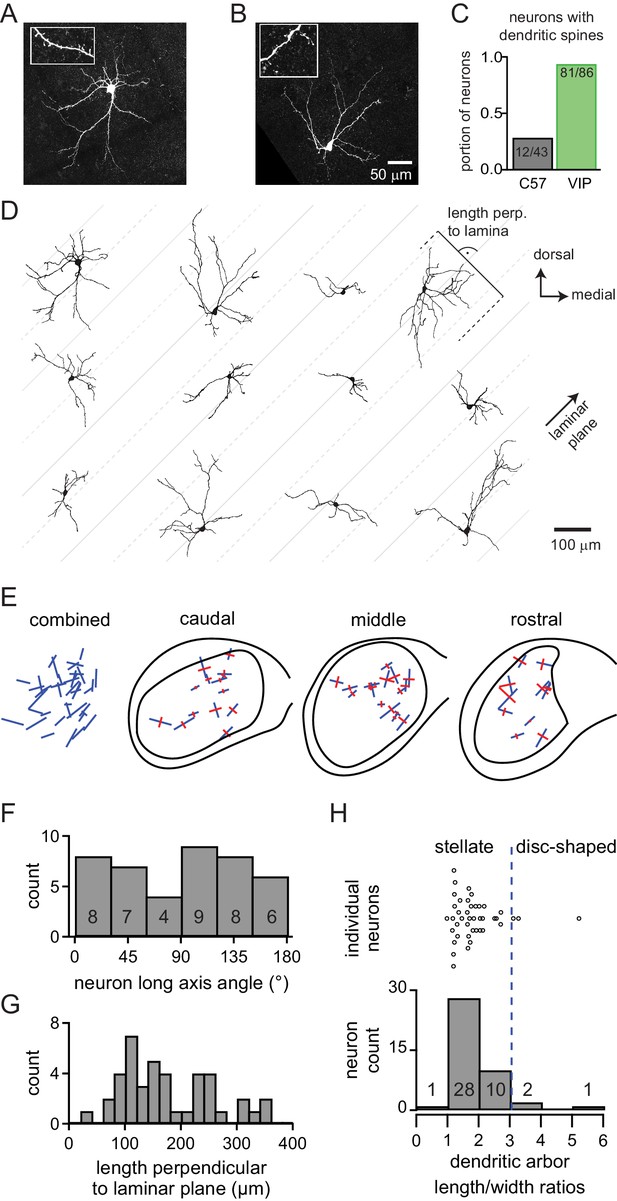
VIP neurons in the ICc are a class of stellate cells and most VIP neurons have dendritic spines.
(A, B) Maximum-intensity projections of confocal z-stacks showing streptavidin-Alexa Fluor-stained VIP neurons from the ICc. Insets: enlarged views of dendritic segments show dendritic spines. (C) 94% of VIP neurons across all IC subdivisions had spiny dendrites vs 28% of neurons from non-targeted recordings in C57BL/6J animals. (D) Representative reconstructions of the morphology of 12 VIP neurons from the ICc. Neurons are oriented as if in the left ICc. Gray lines were drawn at a 45° angle to illustrate the general orientation of the laminae. Solid gray lines are spaced 200 µm apart, dashed lines and solid lines are spaced 100 µm apart. (E) Orientation of the dendritic fields of VIP neurons from the ICc. Combined: Orientation of all reconstructed VIP neurons from the ICc (n = 42). Blue lines denote the orientation of the longest axis (first principal direction) found for each neuron using 2D PCA. Caudal, middle, rostral: Orientation of dendritic fields separated according to position along the rostro-caudal axis of the ICc. Blue lines show longest axis, perpendicular red lines show second longest axis (second principal direction) of each neuron as defined by 2D PCA. (F) Angular orientation of the long axis for every reconstructed VIP neuron within the ICc. Angles indicate counter-clockwise rotation relative to the medial-lateral (horizontal) axis. (G) Spread of the dendritic arbors of ICc VIP neurons measured perpendicular to a predicted 45° isofrequency plane. The dendrites of 83% of VIP neurons extended more than 100 µm across the laminar plane. (H) Dendritic arbor length to width ratio for all reconstructed VIP neurons from the ICc (n = 42). 93% of VIP neurons had a length to width ratio <3, indicating that they are stellate cells. The orientation of length and width axes was determined using 3D PCA.
-
Figure 5—source data 1
Morphometric analysis of VIP neurons in the ICc.
- https://doi.org/10.7554/eLife.43770.013
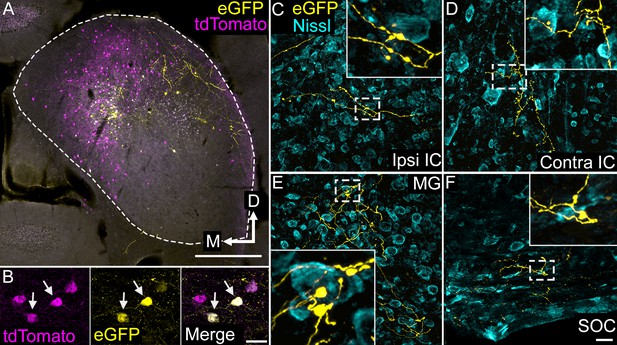
VIP neurons project to multiple local and long-range targets.
(A) Photomicrograph of an AAV deposit site in the right IC. AAV-infected, VIP-expressing cells are labeled with eGFP (yellow), while all VIP-expressing cells are labeled with tdTomato (magenta). Cells expressing both fluorescent proteins appear white. Scale = 500 µm. (B) High magnification photomicrographs showing labeled cells in the AAV deposit site. The field shows four tdTomato-expressing cells (magenta), two of which (white arrows) were also AAV-infected and expressed eGFP (yellow). Scale = 20 µm. (C–F) High magnification photomicrographs showing eGFP-labeled collicular axons (yellow) terminating in the ipsilateral IC (C), the contralateral IC (D), the medial geniculate body (E), or the ventral nucleus of the trapezoid body in the superior olivary complex (F) after an AAV injection in the IC. The white dashed box in each image identifies an area enlarged in the inset to show details of labeled axons and boutons. A fluorescent Nissl stain (cyan) shows that boutons are located in close association with cell bodies as well as in the intervening neuropil. Scale = 20 µm.
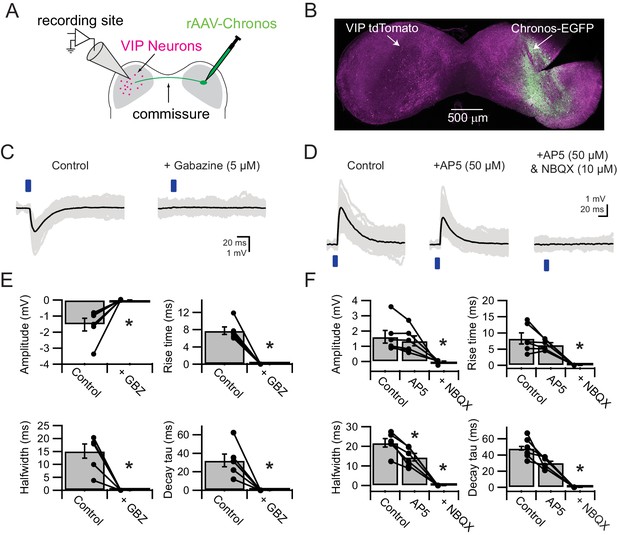
VIP neurons in the ICc receive excitatory and inhibitory synaptic input from the contralateral IC.
(A) Experimental setup. An AAV encoding Chronos-GFP was injected into the right IC. Three weeks later, light-evoked postsynaptic potentials were recorded from VIP neurons in the left ICc. (B) Image of a coronal slice of the IC. Injection sites and Chronos expression were validated through Chronos-GFP fluorescence. (C) Optogenetically-evoked IPSPs recorded from VIP Neurons in the ICc contralateral to the AAV injection site. IPSPs were evoked by 2–5 ms blue light flashes (left), while EPSPs were blocked with NBQX and AP5. IPSPs were abolished by gabazine (right). (D) Optogenetically-evoked EPSPs recorded from VIP neurons in the ICc contralateral to the AAV injection site. EPSPs were evoked by 2–5 ms blue light flashes (left), while IPSPs were blocked with strychnine and gabazine. Wash-in of AP5 significantly reduced the halfwidth and decay time constant of light-evoked EPSPs (middle). Wash-in of NBQX abolished the remaining EPSP (right). (E) Population data showing amplitude and kinetics of optogenetically-evoked IPSPs. (F) Population data showing amplitude and kinetics of optogenetically-evoked EPSPs. The significant reduction of EPSP halfwidth by AP5 indicates that NMDA receptor activation prolonged EPSP duration.
-
Figure 7—source data 1
EPSP and IPSP analysis of commissural inputs to VIP neurons.
- https://doi.org/10.7554/eLife.43770.016
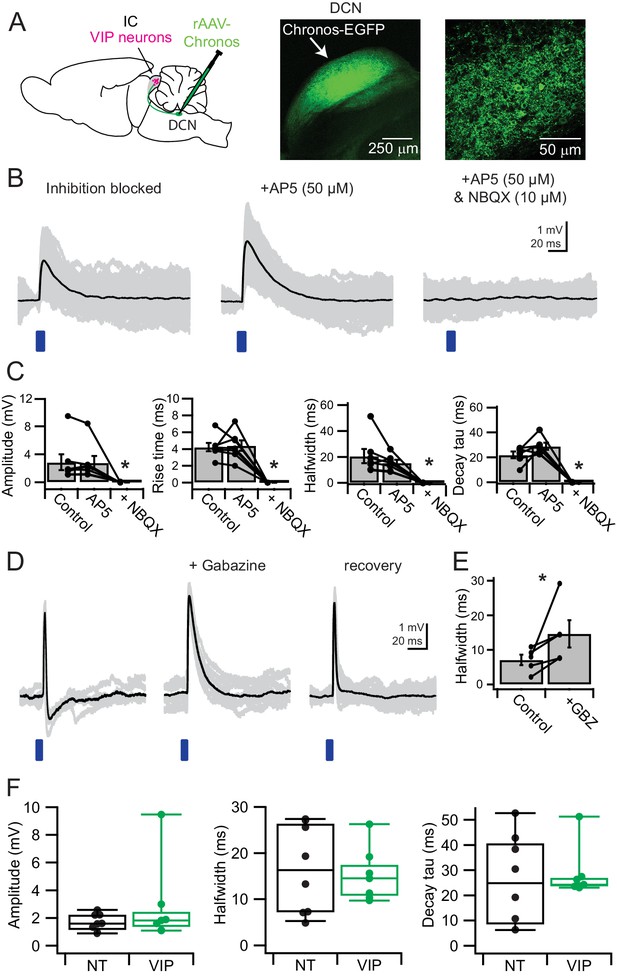
VIP neurons in the ICc receive direct synaptic input from the DCN and feedforward inhibition driven by DCN afferents.
(A) Experimental setup. An AAV encoding Chronos-GFP was injected into the right DCN. For every experiment, the injection site and Chronos-GFP expression were confirmed through GFP fluorescence. Current clamp recordings were made from VIP neurons in the ICc contralateral to the injection site. (B) With inhibition blocked by gabazine and strychnine, 2–5 ms blue light flashes evoked EPSPs (left). AP5 did not significantly reduce EPSP halfwidth or decay time constant. Subsequent addition of NBQX abolished the EPSP. (C) Population data showing amplitude and kinetics for EPSPs elicited by activation of DCN synapses onto VIP neurons in the ICc. The absence of a significant effect of AP5 indicates that NMDA receptors did not make a significant contribution to EPSPs. (D) In several recordings made in the absence of inhibitory blockers, EPSP duration was limited through GABAergic feedforward inhibition (left; n = 5). In these instances, gabazine wash-in increased EPSP halfwidth to values similar to those in (B). (E) Population data for feedforward inhibition to VIP neurons. Washing in gabazine increased EPSP halfwidth in 5 out of 5 tested connections. (F) Population data comparing amplitude, halfwidth, and decay time constant of EPSPs from DCN inputs recorded in VIP neurons (VIP) and non-VIP neurons (NT, non-targeted recording). Halfwidth and decay time constants in VIP neurons showed a trend to cluster more tightly that in non-targeted recordings.
-
Figure 8—source data 1
Analysis of EPSPs from DCN inputs in VIP neurons and non-targeted recordings.
- https://doi.org/10.7554/eLife.43770.018
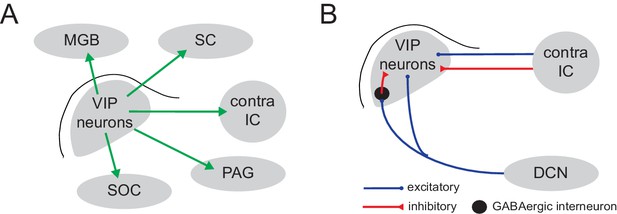
Summary of inputs and projection targets of VIP neurons in mouse IC.
(A) Summary of the major projection targets of VIP neurons identified by axonal tract tracing: MGB (medial geniculate body), SC (superior colliculus), contralateral IC, PAG (periaqueductal gray), SOC (superior olivary complex). (B) Summary of the sources of input to VIP neurons identified by CRACM experiments.
Tables
VIP neurons are glutamatergic.
Across three mice, an average of 1.3% of tdTomato+ neurons were labeled with an antibody against GAD67.
| Animal | Slice # | # tdTomato+ | # GAD67+ | # co-labeled | % tdTomato+co-labeled |
|---|---|---|---|---|---|
| P58 female, #1 | 1 (caudal) | 210 | 184 | 3 | 1.4 |
| 2 (middle) | 172 | 65 | 2 | 1.2 | |
| Total | 382 | 249 | 5 | 1.3 | |
| P58 male | 1 (caudal) | 151 | 152 | 2 | 1.3 |
| 2 (middle) | 46 | 212 | 2 | 4.3 | |
| Total | 197 | 364 | 4 | 2.0 | |
| P58 female, #2 | 1 (caudal) | 161 | 137 | 0 | 0.0 |
| 2 (middle) | 53 | 187 | 1 | 1.9 | |
| Total | 214 | 324 | 1 | 0.5 | |
| Grand total | 793 | 937 | 10 | 1.3 | |
| Average across three mice (mean ± SD) | 1.3 ± 0.8% | ||||
VIP neurons represent 3.5% of ICc neurons, 1.5% of IC shell neurons, and are present at a higher density in the caudal ICc and IC shell.
Table shows results from stereological analysis of the percentage of neurons (NeuN+) in the ICc and IC shell (ICd + IClc) that express tdTomato in VIP-IRES-Cre x Ai14 mice. Values indicate mean ± SEM, (#tdTomato+ neurons / #NeuN+ neurons), and number of systematic random samples analyzed from each slice.
| ICc | ||||
|---|---|---|---|---|
| Coronal slice | P54 male 1 | P54 male 2 | Per slice plane | Grand average |
| Caudal | 3.1 ± 0.9% (12/503) five samples | 8.4 ± 1.2% (26/338) four samples | 5.8 ± 2.7% (38/841) | |
| Middle | 2.4 ± 0.8% (20/741) eight samples | 3.9 ± 0.9% (44/1173) eight samples | 3.2 ± 0.7% (64/1914) | |
| Rostral | 1.9 ± 0.6% (21/929) eight samples | 1.2 ± 0.4% (12/1024) seven samples | 1.5 ± 0.4% (33/1953) | |
| Per mouse | 2.5 ± 0.3% (53/2173) | 4.5 ± 2.1% (82/2535) | 3.5 ± 1.0% (135/4708) | |
| IC shell | ||||
| Coronal slice | P54 male 1 | P54 male 2 | Per slice plane | Grand average |
| Caudal | 2.9 ± 0.8% (35/1092) 10 samples | 1.9 ± 0.8% (9/615) five samples | 2.4 ± 0.5% (44/1707) | |
| Middle | 0.9 ± 0.6% (4/534) six samples | 0.9 ± 0.6% (10/944) eight samples | 0.9 ± 0.0% (14/1478) | |
| Rostral | 1.2 ± 0.4% (10/842) eight samples | 1.2 ± 0.6% (5/569) four samples | 1.2 ± 0.0% (15/1411) | |
| Per mouse | 1.6 ± 0.6% (49/2468) | 1.3 ± 0.3% (24/2128) | 1.5 ± 0.2% (73/4596) | |
-
Table 2—source data 1
Percentages of VIP neurons in ICc and IC shell.
- https://doi.org/10.7554/eLife.43770.007
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6J | The Jackson Laboratory | JAX:000664 | |
| Genetic reagent (Mus musculus) | VIP-IRES-Cre | The Jackson Laboratory | JAX:010908 | |
| Genetic reagent (Mus musculus) | Ai14 | The Jackson Laboratory | JAX:007914 | |
| Antibody | anti-GAD67 (mouse monoclonal) | Millipore | RRID:AB_2278725 Cat#:MAB5406 | IHC (1:1000) |
| Antibody | anti-NeuN (rabbit polyclonal) | Millipore | RRID:AB_10807945 Cat#:ABN78 | IHC (1:500) |
| Antibody | anti-bNOS (mouse monoclonal) | Sigma-Aldrich | RRID:AB_260754 Cat#:N2280 | IHC (1:1000) |
| Antibody | anti-mouse IgG Alexa Fluor 488 (donkey polyclonal) | ThermoFisher | RRID:AB_141607 Cat#:A-21202 | IHC (1:500) |
| Antibody | anti-rabbit IgG Alexa Fluor 488 (donkey polyclonal) | ThermoFisher | RRID:AB_2535792 Cat#:A-21206 | IHC (1:500) |
| Recombinant DNA reagent | AAV1.Syn.Chronos-GFP.WPRE.bGH | University of Pennsylvania Vector Core/Addgene | Addgene:59170-AAV1 RRID:Addgene_59170 | http://n2t.net/addgene:59170 |
| Recombinant DNA reagent | AAV1.CAG.FLEX.eGFP.WPRE.bGH | University of Pennsylvania Vector Core/Addgene | Addgene:51502-AAV1 RRID:Addgene_51502 | http://n2t.net/addgene:51502 |
| Chemical compound, drug | gabazine | Hello Bio | Cat#:HB0901 | also called SR95531 hydrobromide |
| Chemical compound, drug | strychnine hydrochloride | Sigma-Aldrich | Cat#:S8753 | |
| Chemical compound, drug | D-AP5 | Hello Bio | Cat#:HB0225 | |
| Chemical compound, drug | NBQX disodium salt | Hello Bio | Cat#:HB0443 | |
| Software, algorithm | Igor Pro 7 and 8 | Wavemetrics | RRID:SCR_000325 | |
| Software, algorithm | MATLAB R2018a and R2018b | Mathworks | RRID:SCR_001622 | |
| Software, algorithm | Neurolucida | MBF Bioscience | RRID:SCR_001775 | |
| Software, algorithm | Neurolucida 360 | MBF Bioscience | RRID:SCR_016788 |
Stereotaxic coordinates for virus injections. All coordinates are relative to the lambda suture.
| Location | X coordinate (caudal) | Y coordinate (lateral) | Z coordinates (depth) |
|---|---|---|---|
| Right IC penetration 1 (CRACM) | -900 µm | 1000 µm | 2250 - 1500 µm, 250 µm interval |
| Right IC penetration 2 (CRACM) | -900 µm | 1250 µm | 2250 - 1750 µm, 250 µm interval |
| Right IC penetration 1 (axonal tracing) | -900 µm | 1000 µm | 1850 µm, 2000 µm |
| Right DCN | -1325 µm | 2150 µm | 4750 µm, 4550 µm |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43770.021





