Evolution of limb development in cephalopod mollusks
Figures
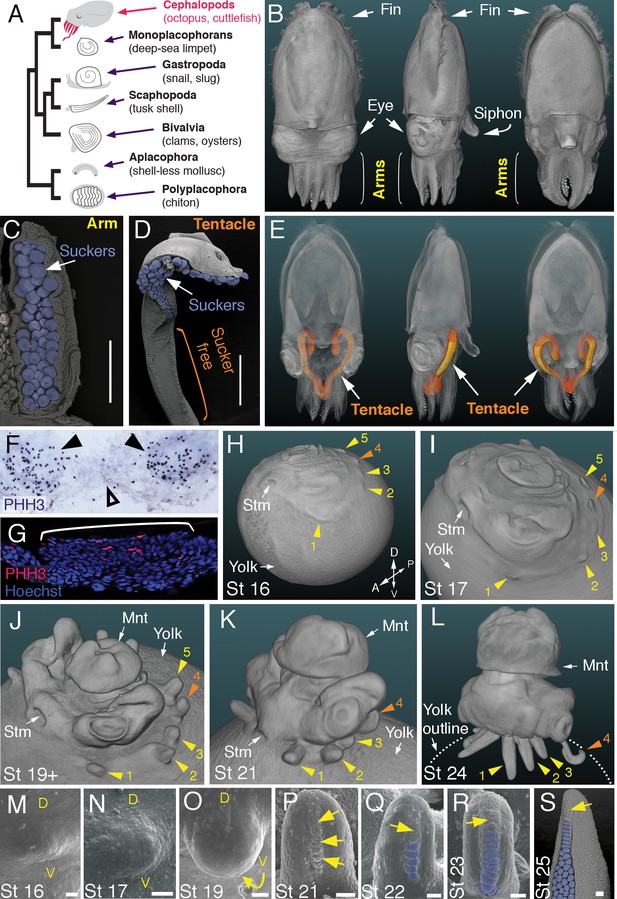
Development of arms and tentacles in the cuttlefish, Sepia officinalis.
(A) Phylogenetic relationships of Mollusca based on phylogenomic data (Smith et al., 2011) illustrating the unique morphology of the cephalopod body plan compared to other mollusks. (B) OPT reconstruction of a cuttlefish hatchling showing positions of the limbs; only arms are visible (see also Figure 1—video 1). (C to D), SEM of the ventral side of a cuttlefish arm (C) and tentacle (D). Suckers are pseudocolored blue. Note distal restriction of suckers in tentacle relative to arm. (E) OPT reconstruction illustrating the internally retracted tentacles. Specimens are same as in (B), but here the tentacles are displayed in orange and the rest of the tissue is rendered translucent (see also Figure 1—video 2). (F and G) Phospho-histone H3 (PHH3) immunostaining at stage 15 shows localized clusters of proliferating cells at the onset of limb development (black arrowheads) but little proliferation in the interlimb region (open arrowhead). (F) PHH3 detection by colorimetric reaction with DAB in a whole mount. (G) PHH3 immunofluorescence (red) on limb cryosection (white bracket). Cell nuclei (blue) are labeled by Hoechst staining. (H-L) OPT reconstructions of cuttlefish embryos at stages 16 to 24. Cuttlefishes have five bilaterally symmetric limb pairs (ten limbs; eighth arms and two tentacles). Numbered arrowheads mark all five limbs/limb buds on the left side of each embryo. The left tentacle differentiates from position number four (orange arrowhead), whereas arms form from limb buds at the other positions (yellow arrowheads). See also Figure 1—videos 3—5. A, anterior; P, posterior; D, dorsal; V, ventral; Stm, stomodeum; Mnt, mantle. (M to O) SEM during early stages of cuttlefish limb development (stages 16 to 19). Morphogenesis of the limb is first observed as a slight swelling (M) that transforms into a limb bud (O) as proximodistal outgrowth progresses. D, dorsal; V, ventral. (P–S), SEM at later stages of cuttlefish limb development (stages 21 to 25) showing the formation of sucker buds on the ventral surface of a developing limb. A primordial sucker band (yellow arrows) is observed along the ventral midline of a stage 21 limb bud (P). At later stages, the band cleaves superficially from the proximal end to form the sucker buds (pseudocolored blue in Q to S). Scale bars: 0.5 mm (C and D) and 100 μm (M to S).
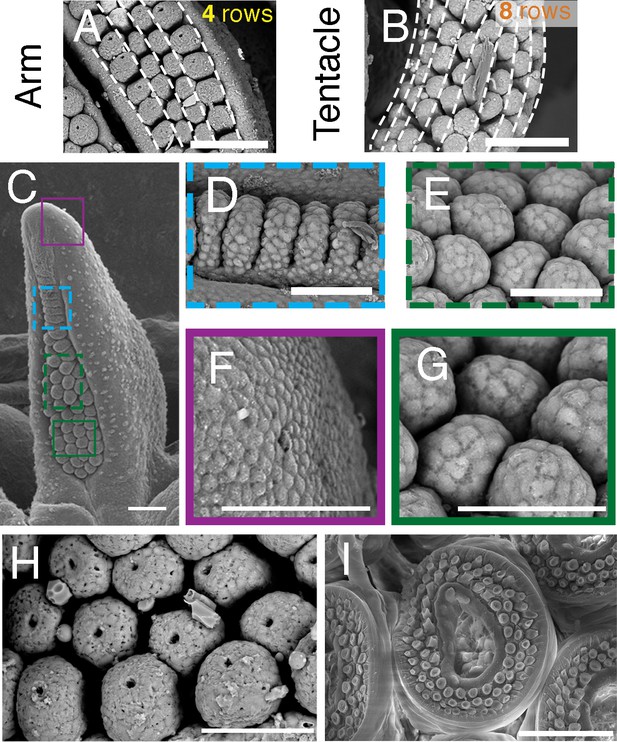
Sucker morphogenesis.
All panels show scanning electron micrographs; scale bars, 100 μm. (A and B) Sucker buds are arranged in parallel rows along the anteroposterior axis, with four rows in arms (A) and eight rows in tentacles (B). (C-G) Sucker formation progresses from distal to proximal. Colored squares in C are shown at higher magnification in D-G. Superficial cleavage of the proximal side of the primordial sucker band in (D) and segregation of the recently formed sucker buds in (E). Early sucker bud cells (G) form a dome-shaped outline compared to the rather flattened morphology of the non-sucker forming surface epithelium (F). (H and I) Higher magnification of sucker buds in a cuttlefish hatchling showing that sucker differentiation is not yet complete in hatchlings (H) compared to the differentiated suckers found in more mature individuals (I), which indicates that a substantial portion of sucker development occurs during post hatchling development.
OPT 3D reconstruction showing cuttlefish hatchling morphology.
https://doi.org/10.7554/eLife.43828.005OPT 3D reconstruction showing the internal location of the tentacles in a cuttlefish hatchling.
Tentacles in orange, other parts of the body in gray (partially translucent).
OPT 3D reconstruction showing morphology of a cuttlefish embryo at stage 17.
Embryo is positioned on top of the yolk. The early limb buds (eight arm buds and two tentacle buds) can be seen around the margin of the embryo.
OPT 3D reconstruction showing morphology of a cuttlefish embryo at late stage 19.
https://doi.org/10.7554/eLife.43828.008OPT 3D reconstruction showing the morphology of a cuttlefish embryo at stage 24.
https://doi.org/10.7554/eLife.43828.009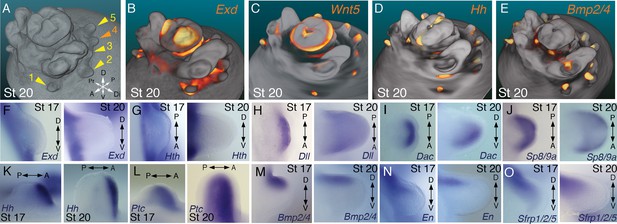
Molecular regionalization of proximodistal, anteroposterior, and dorsovental axes during cephalopod limb development.
(A) OPT reconstruction of cuttlefish embryo at stage 20 showing all five limb buds on the left side of the embryo (arms, yellow arrowheads; tentacle, orange arrowhead). (B to E) OPT reconstructions showing four representative genes with polarized expression patterns along major axes of limb buds (gene expression indicated by orange/yellow). Proximodistally polarized expression of Exd (B) and Wnt5 (C). Anteroposteriorly polarized expression of Hh (D), dorsoventrally polarized expression of Bmp2/4 (E). (F to O), In situ hybridizations of cuttlefish limb buds at stage 17 (left) and stage 20 (right) showing polarized patterns of expression along the proximodistal axis for Exd (F), Hth (G), Dll (H), Dac (I) and Sp8/9a (J); the anteroposterior axis for Hh (K) and Ptc (L); and the dorsoventral axis for Bmp2/4 (M), En (N) and Sfrp1/2/5 (O). A, anterior; P, posterior; D, dorsal; V, ventral; Di, distal; Pr, proximal.
-
Figure 2—source data 1
Source data for molecular phylogenetic analyses.
- https://doi.org/10.7554/eLife.43828.021
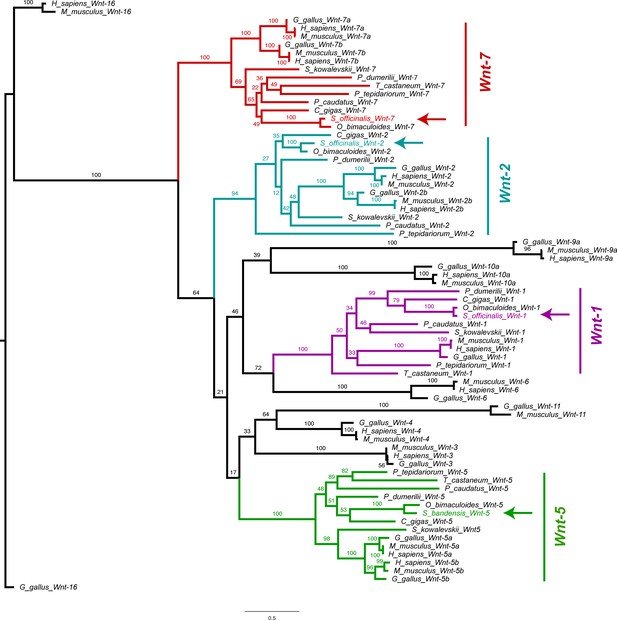
Wnt phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Wnt family ligands isolated in this study. Arrows mark the phylogenetic placement of 4 cuttlefish Wnt sequences.
Pan/Tcf phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Pan/Tcf transcription factors isolated in this study. The arrow marks the phylogenetic placement of one cuttlefish Tcf sequences.
Dac/Dach phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Dac/Dach transcription factors isolated in this study. The arrow marks the phylogenetic placement of one cuttlefish Dac sequences.
Sp phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Sp family of transcription factors isolated in this study. The arrow marks the phylogenetic placement of one cuttlefish Sp sequence.
Homeodomain transcription factor phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of homeodomain transcription factors isolated in this study. Arrows mark the phylogenetic placement of 4 cuttlefish homeodomain sequences.
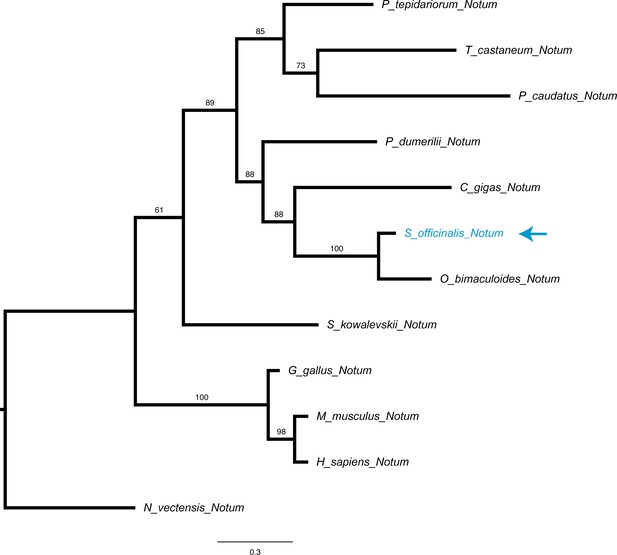
Notum phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Notum Wnt inhibitors isolated in this study. The arrow marks the phylogenetic placement of one cuttlefish Notum sequence.
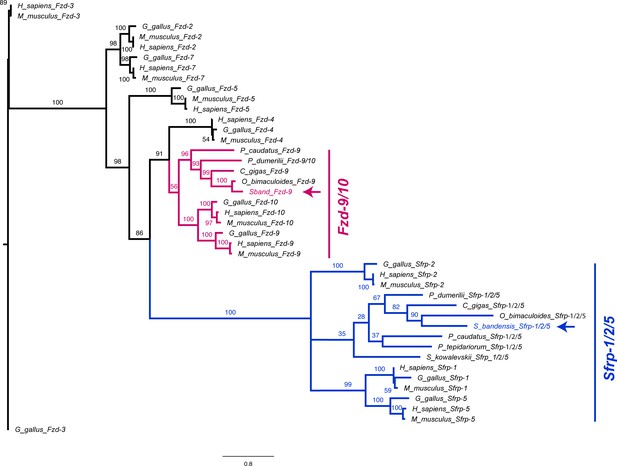
Frizzled/Sfrp phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Frizzled Wnt co-receptors and Sfrp extracellular Wnt repressors isolated in this study. Arrows mark the phylogenetic placement of one cuttlefish Frizzled and one Sfrp.
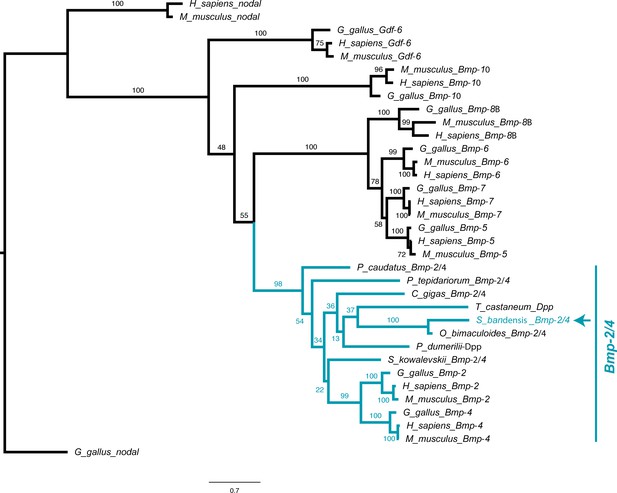
Tgfβ phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Tgfβ family ligands isolated in this study. The arrow marks the phylogenetic placement of cuttlefish Bmp2/4.
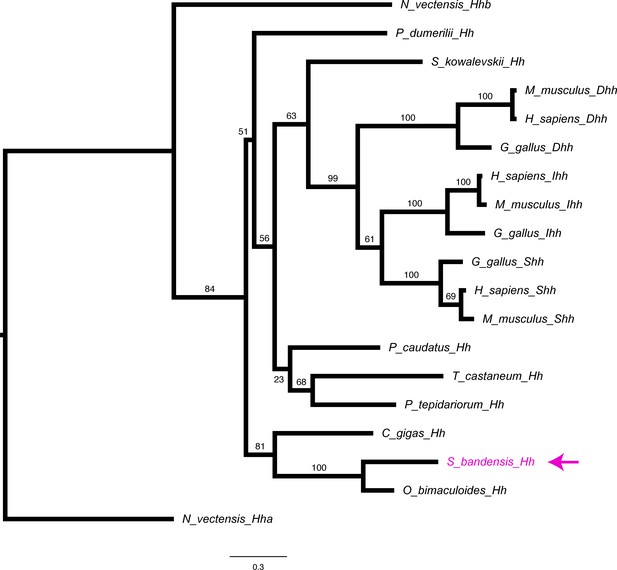
Hedgehog phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Hh ligand previously isolated (Tarazona et al., 2016) in this study. The arrow marks the phylogenetic placement of cuttlefish Hh sequence.
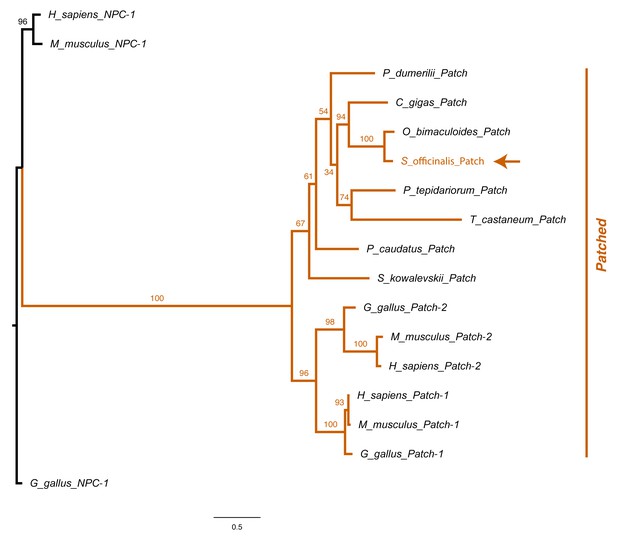
Patched phylogeny.
Molecular phylogenetic reconstruction using maximum likelihood implemented in RAxML of Hedgehog Patched receptors isolated in this study. The arrow marks the phylogenetic placement of cuttlefish Patched.
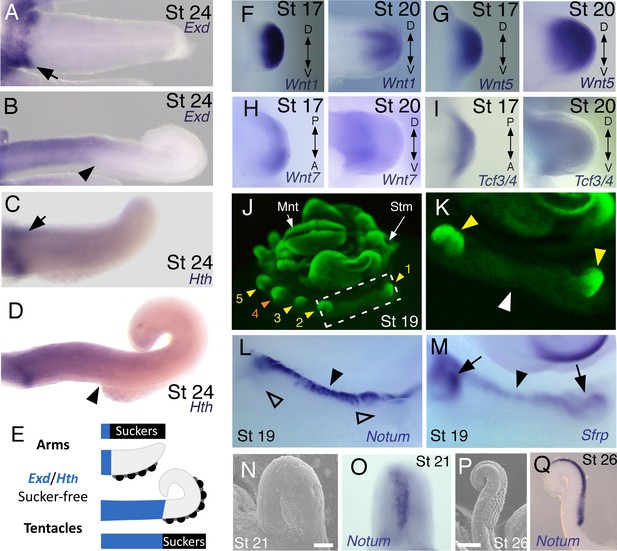
Expression of proximal identity genes Exd and Hth in arms and tentacles corresponds with distribution of suckers; Wnt signaling repressors are dorsally restricted.
(A and B) Compared to arms (A), tentacles (B) show a distally expanded domain of Exd expression in the proximal region of the limb. (C and D) A similar pattern of expression is detected for Hth during arm (C) and tentacle (D) development. Distal boundary of Exd and Hth expression marked by black arrowheads in (A to D). (E) Expanded expression of proximal identity genes correlates with the expanded sucker-free domain seen in tentacles compared to arms. (F and H) The Wnt ligands Wnt1, Wnt5 and Wnt7 show a distally restricted expression but no dorsoventral polarization at stages 17 and 20. (I) The Wnt signaling transcription factor Tcf3/4 is also distally restricted but shows no dorsoventral polarization at stages 17 and 20. (J and K) Fluorescent nuclear stain SYBR Safe highlights limb buds (yellow arrowheads). Boxed region in (J) is enlarged in (K); white arrowhead marks interlimb region. (L and M) The Wnt ligand repressors Notum and Sfrp1/2/5 are expressed in the dorsal interlimb region (black arrowhead in L and M; compare with K). Sfrp1/2/5 expression expands into the dorsal limb bud (black arrows in M) in stage 19 embryos, whereas Notum stays dorsal but proximally restricted (open arrowheads mark the limb buds in L). (N and O) The earliest sign of sucker formation can be detected by SEM as a slight swelling (N) and by Notum expression (O) on the ventral side of stage 21 limb buds. (P and Q) Expression of Notum is maintained through later stages of sucker morphogenesis, as seen in stage 26 tentacles (lateral views).
-
Figure 3—source data 1
Sequence similarities of Sepia bandensis clones.
- https://doi.org/10.7554/eLife.43828.025
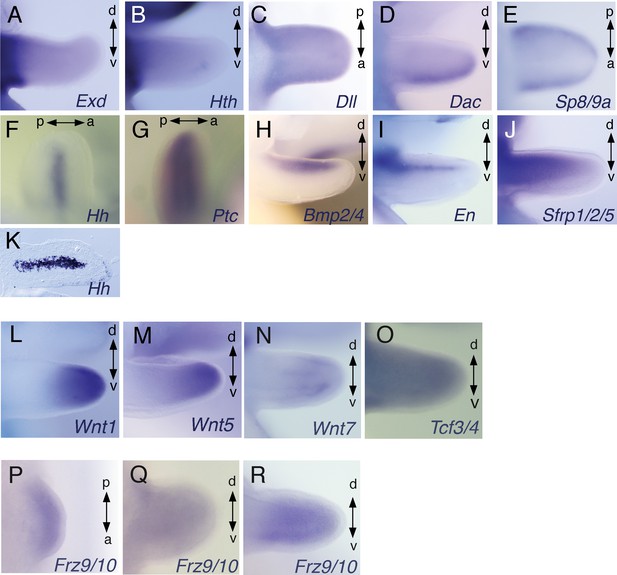
Expression of developmental control genes in cuttlefish limb buds.
(A to O) In situ hybridizations showing Wnt1, Wnt5, Wnt7, Tcf, Exd, Hth, Dll, Dac, Sp8, Hh, Ptc, Bmp2/4, En and Sfrp1/2/5 in stage 21 embryos. Hh expression in stage 21 limb buds, detected by in situ hybridizations in whole mount (F) and cryosections (K) showing the central expression in the brachial nerve cell precursors. (P to R) In situ hybridizations of Frz9/10 at stages 17 (P and S), 20 (Q and T) and 21 (R and U). A, anterior; P, posterior; D, dorsal; V, ventral.
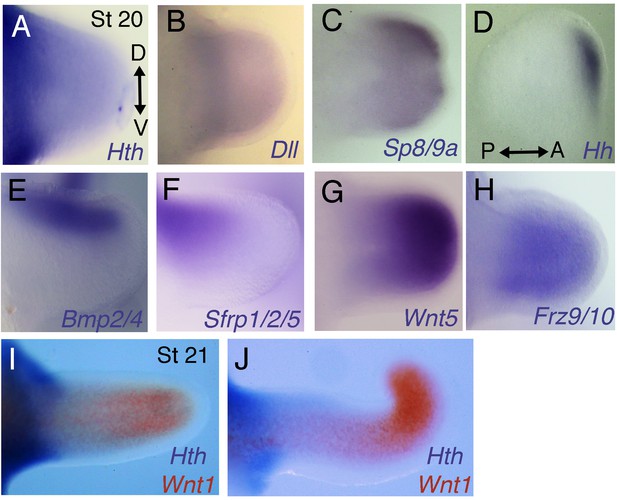
Analysis of gene expression in the arms and tentacles of Sepia bandensis embryos.
S. bandensis probes that were used on S. officinalis embryos were validated by species-specific hybridization using S. bandensis embryos. Each probe yielded identical patterns of expression in the limb buds of the two Sepia species. (A–C) Stage 20 S. bandensis limb buds show proximodistally regionalized expression of Hth, Dll and Sp8/9a. The Hth domain (A) is proximal to the Dll (B) and Sp8/9a (C) expression domains. (D) Hh is restricted to the anterior margin of the limb bud of S. bandensis. (E, F), Dorsal expression of Bmp2/4 (E) and Sfrp1/2/5 (F) in stage 20 S. bandensis limb buds. (G, H) Wnt5 (G) and Frz9/10 (H) are expressed in the distal but are undetectable in the proximal regions of S. bandensis limb buds. (I, J) Two-color in situ hybridizations show that proximally restricted expression of Hth (purple) and distally restricted expression of Wnt1 (red) are retained during S. bandensis limb outgrowth. Stage 21 arm (I) and tentacle (J). D, dorsal; V, ventral; P, posterior; A, anterior. All limb buds oriented as in (A) except for (D), which is oriented according to the axes shown.
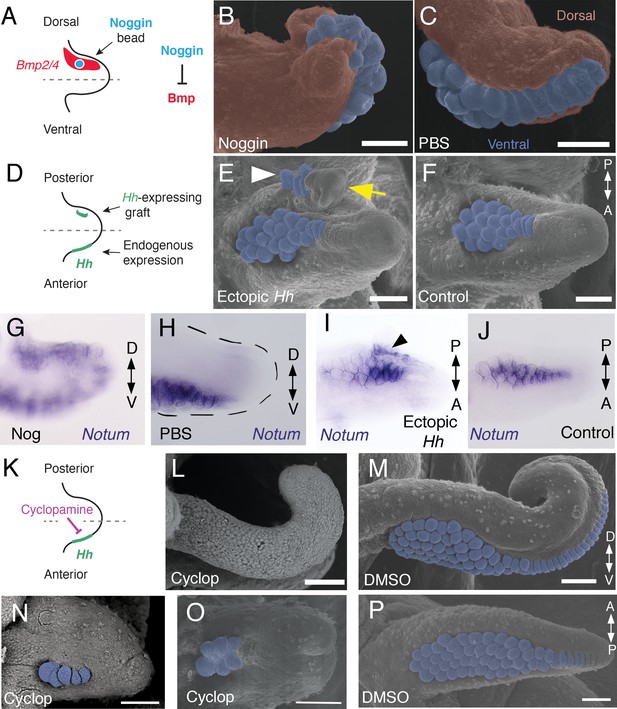
Bmp signaling controls dorsoventral patterning and Hh signaling regulates anteroposterior patterning of cuttlefish limbs.
(A to C) Implantation of carrier beads loaded with the Bmp inhibitor Noggin (A) results in formation of ectopic sucker buds (n = 8/12) on the dorsal surface of the limb (B), whereas PBS control beads (n = 15/15) result in normal development of the dorsal limb (pseudocolored red) restriction of suckers (pseudocolored blue) to the vental side of the limb (C). (D-F), Hh-expressing cells (taken from the funnel of a stage 24 donor embryo) grafted to the posterior side of a stage 17 cuttlefish limb bud (D) generates a posterior mirror-image limb duplication (n = 7/12; yellow arrow in E), whereas no duplication (n = 8/8) results when control (Hh-negative) cells are grafted to the same position (F). Sucker buds are pseudocolored blue in (E and F); sucker buds in duplicated limb marked with a white arrowhead. (G and H) Noggin beads induce ectopic expression of Notum on the dorsal side of the limb (G). Limbs receiving control PBS beads show normal expression of Notum ventrally (H). (I and J) Graft of Hh-expressing tissue to the posterior side of the limb induces ectopic domain of Notum prior to duplication of the limb (I; black arrowhead). Note the two separate domains of Notum expression in I compared to a single Notum expression domain in the limb with the Hh-negative control graft (J). (K to P) Transitory repression of Hh signaling by cyclopamine (K) during early stages of limb development disrupts the anteroposterior distribution of sucker bud rows (L, N, O). Cyclopamine-treated limbs showing complete loss of suckers in tentacles (L) and reduction in the number of sucker bud rows in arms (N and O). Control embryos treated with vehicle only (DMSO) develop the normal number of sucker bud rows in tentacles (M) and arms (P). Sucker buds are pseudocolored blue in (B, E, F, and M- P). Scale bars 100 μm.
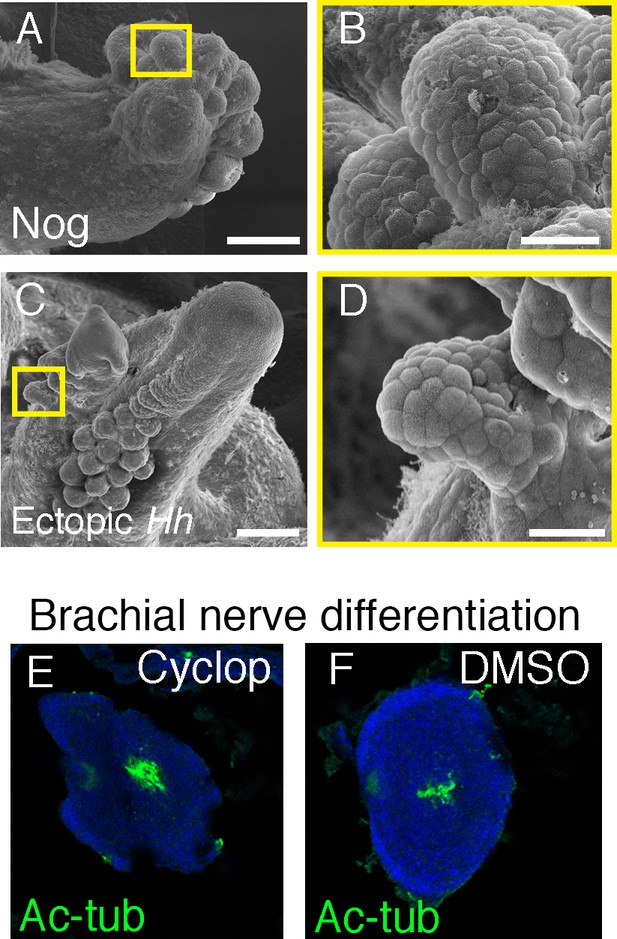
Sucker development after manipulations of Bmp and Hh signaling pathways.
(A) SEM of Noggin-treated limbs shows dorsal ectopic sucker buds (A) as seen in Figure 4B. (B) Higher magnification of the region inside the yellow square in (A) showing the superficial dome-shape morphology of sucker bud cells. (C) SEM of posterior mirror-image duplicated limb caused by graft of Hh-expressing tissue, as seen in (Figure 4e). (D) Higher magnification of the region inside the yellow square in (C) shows the superficial dome-shape morphology of sucker bud cells. (E and F) Differentiation of brachial nerves in cyclopamine-treated (E) and DMSO control (F) embryos revealed by acetylated tubulin in the center of the limbs. Scale bars, 100 μm.
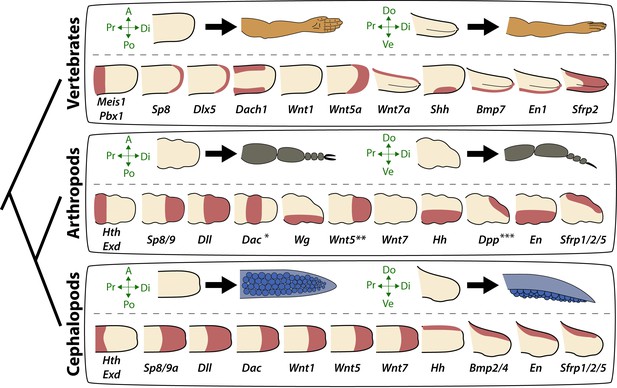
Molecular patterning of the anteroposterior, proximodistal, and dorsoventral axes of developing limbs in vertebrates, arthropods, and cephalopods.
For each lineage, the top row shows schematic representations of a generalized limb bud and an adult limb in two different orientations. Axes are indicated to the left of each limb bud (A, anterior; Po, posterior; Pr, proximal; Di, distal; Do, dorsal; Ve, ventral) Bottom rows show limb buds with gene expression domains pink color). Vertebrate gene expression based on mouse limb development. Arthropod gene expression is a compound reconstruction from chelicerate, myriapod, and hexapod limb development in order to consolidate a complete set of pro-orthologous genes comparable to vertebrate and cephalopod lineages. Cephalopod gene expression is based on findings in this study from the cuttlefish Sepia officinalis and Sepia bandensis. The figure illustrates the conserved and divergent expression patterns of homologous (orthologous) genes, some of which share equivalent roles in patterning the limb axes. The proximodistal axis displays conserved expression of transcription factors at opposite ends; Hth (pro-ortholog of vertebrate Meis genes) and Exd (pro-ortholog of vertebrate Pbx genes) are restricted proximally, whereas Dll (pro-ortholog of vertebrate Dlx genes), Wnt5 (pro-ortholog of Wnt5a) and Sp8/9 (pro-ortholog of vertebrate Sp8 and Sp9 genes, known as Sp6-9 in some arthropods) show distally restricted expression. The typical expression pattern of Dac seen in arthropods (between proximally and distally restricted genes) is not strictly conserved in vertebrates (Dac is the pro-ortholog of vertebrate Dach genes) or cephalopods. However, Dac expression in non-locomotory arthropod appendages (e.g., mandibles) is distally restricted, resembling cephalopod Dac expression (Donoughe and Extavour, 2016). Expression patterns of the diverse family of Wnt genes shows interesting variation. Although, some members of the family show variation in their expression pattern (Wnt1 and Wnt7), there is a general pattern of distal restriction of Wnt expression (represented here by Wnt5, but also seen in many other Wnt ligands) in the three lineages. At the level of individual Wnt members, Wg (pro-ortholog of vertebrate Wnt1) is restricted ventrally in arthropods but not in vertebrates or cephalopods, and Wnt7a (arthropod and cephalopod Wnt7 genes are pro-orthologs of vertebrate Wnt7a) is restricted dorsally in vertebrates but not in arthropods or cephalopods. Restricted expression of Wnt ligands either dorsally or ventrally has not been reported in cephalopods, but the dorsally restricted expression of the Wnt repressor Sfrp1/2/5 suggests a role of polarized Wnt pathway activation in the control of the dorsoventral axis of cephalopod limbs, similar to vertebrates (by dorsal Wnt7a) and arthropods (by ventral Wg). There is a clear restriction of at least one Bmp ligand (vertebrate Bmp7 and cephalopod Bmp2/4; pro-orthologs of arthropod Dpp) and the transcription factor En along the dorsoventral axis in these three lineages. Finally, polarized expression of Hh is conserved in the three lineages (posterior in vertebrates and arthropods, but anterior in cephalopods), which, together with the functional manipulations, indicates conservation of Hh signaling in patterning the anteroposterior limb axis in the three lineages. The asterisk (*) in arthropod Dac indicates that some mouth appendages show a distal expression domain (Donoughe and Extavour, 2016) (Angelini and Kaufman, 2005) more similar to cephalopod Dac limb expression than to Dac expression in arthropod legs. Two asterisks (**) indicate that Wnt5 expression shows variation in arthropods, with a sub-distal expression in chelicerates (Damen, 2002) but distal in hexapods (i.e. flour beetle) (Bolognesi et al., 2008). Three asterisks (***) indicate that Dpp shows variation in its expression domain in arthropods, with some hexapods and chelicerates showing a distal expression domain, whereas in Myriapods and other hexapods it is dorsally restricted, as depicted here (Angelini and Kaufman, 2005). Schematized gene expression domains for vertebrates and arthropods are from the following sources. Mouse gene expression: Meis1 (González-Lázaro et al., 2014), Pbx1 (Capellini et al., 2006), Sp8 (Kawakami et al., 2004), Dlx5 (Vieux-Rochas et al., 2013), Dach1 (Salsi et al., 2008), Wnt1, Wnt5a, Wnt7a, Sfrp2 (Witte et al., 2009), Shh (Riddle et al., 1993), Bmp7 (Choi et al., 2012) and En1 (Loomis et al., 1998). Arthropod expression based on: Chelicerates, Hth, Exd, Dll, Dac (Prpic et al., 2003), Sp8/9 (Königsmann et al., 2017), Wg (Damen, 2002), En (Damen, 2002) and Sfrp1/2/5 (Hogvall et al., 2018); Myriapods, Hth, Exd, Dll, Dac (Prpic and Tautz, 2003), Sp8/9 (Setton and Sharma, 2018), Wg, Dpp (Prpic, 2004), Wnt5, Wnt7, Hh, En (Janssen et al., 2004), Sfrp1/2/5 (Hogvall et al., 2018); Hexapods, flour beetle, Hth, Exd (Prpic et al., 2003), Sp8/9 (Schaeper et al., 2010), Dll (Beermann et al., 2001), Dac (Prpic et al., 2001), Wg, Wnt5 (Bolognesi et al., 2008), Dpp (Sanchez-Salazar et al., 1996), En (Brown et al., 1994); Hexapods, cricket, Hth, Exd, Dll, Dac, Wg, Hh, Dpp, En (Donoughe and Extavour, 2016).
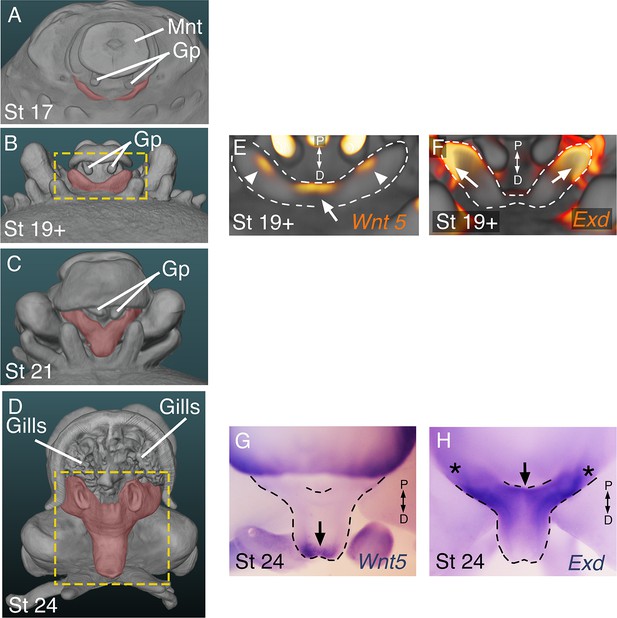
The developing funnel/siphon organ shows limb-like expression patterns of the proximodistal patterning genes Exd and Wnt5.
(A-D) OPT reconstructions of stage 17 to 24 cuttlefish embryos showing the developing funnel/siphon system (pseuodocolored red). (A) Funnel development begins at stage 17 with emergence of two bilaterally symmetrical funnel ridges between the row limb buds (bottom) and the gill primordia (GP), which can be seen adjacent to mantle (Mnt). (B) The funnel ridges fuse medially to form the funnel primordum. Yellow box marks area shown in E and F. (C) A single siphon tube then develops at the distal end of the funnel primordium. (D) By stage 24, the funnel/siphon organ has reached a miniature version of its adult shape and form. At this stage, the mantle covers the proximal region of the funnel. To allow the entire funnel/siphon organ to be seen, we segmented out the funnel and digitally removed part of the mantle using Amira Software. (E–H) Whole mount in situ hybridizations show polarized expression of Wnt5 and Exd in the distal and proximal regions, respectively, of the developing funnel primordium. OPT reconstructions (E, F) and brightfield microscopy (E, G). Wnt5 is expressed distally (arrows) at early and late stages of funnel/siphon development. In addition to the distal medial domain of Wnt5 expression domain at stage 19+ (white arrow in E), two lateral spots of expression can be seen (white arrowheads in E). (F, H) Exd is expressed proximally (arrows) at both stages. Asterisks mark expression of Exd in the proximal ends of the funnel that are situated beneath the mantle.





