Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses
Figures
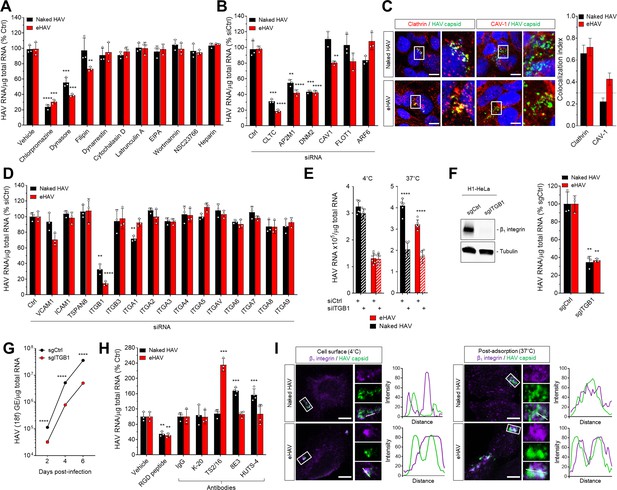
Naked and quasi-enveloped HAV virions undergo clathrin-dependent endocytosis facilitated by β1 integrin.
(A) Effect of endocytic inhibitors on HAV and eHAV entry quantified by RT-PCR (mean ± SD, n = 3 independent experiments). (B) Effect of siRNA-mediated depletion of endocytic regulators on HAV and eHAV entry (mean ± SD, n = 3 independent experiments). See Figure 1—figure supplement 2A for knockdown efficiencies. (C) Confocal micrographs of Huh-7.5 cells immunolabeled with anti-HAV capsid (K24F2) and anti-clathrin or anti-caveolin-1 at one hpi. Scale bar, 10 µm. (D) Effect of siRNA-mediated depletion of integrins and adhesion molecules on HAV and eHAV entry (mean ± SD, n = 3 independent experiments). See Figure 1—figure supplement 2B for knockdown efficiencies. (E) Effect of β1 integrin knockdown on HAV/eHAV attachment to cells 2 hpi at 4˚C (mean ± SD, n = 6 biological replicates from two independent experiments) or on HAV and eHAV uptake 6 hpi at 37˚C (mean ± SD, n = 3 independent experiments). (F) Levels of HAV RNA at 20 hpi in H1-HeLa engineered by CRISPR/Cas9 using a control or a specific ITGB1-targeting sgRNA (mean ± SD, n = 3 biological replicates of a representative experiment). (G) Replication of rapid-replicating HAV (18 f) in sgITGB1 H1-HeLa cells (mean ± SD, n = 3). (H) Effect of an inhibitory RGD-containing peptide or β1 integrin activating antibodies on HAV and eHAV uptake (mean ± SD, n = 3–4 biological replicates of a representative experiment). (I) Confocal micrographs of Huh-7.5 cells incubated at 4˚C or 37˚C for 1 hr and immunolabeled with anti-HAV capsid (K24F2) and anti-β1 integrin. Scale bar, 10 µm. For numeric data plotted in graphs associated with this figure, see Figure 1—source data 1.
-
Figure 1—source data 1
Source data corresponding to Figure 1.
- https://doi.org/10.7554/eLife.43983.008
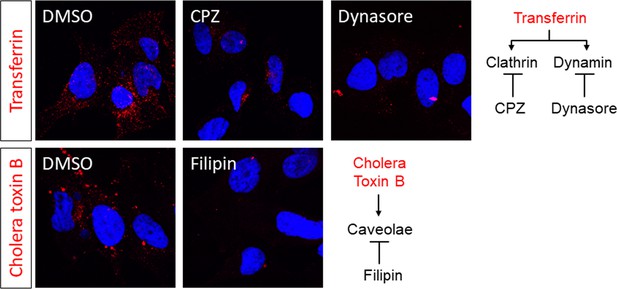
Huh-7.5 cells were incubated with DMEM supplemented with the indicated inhibitors for 1 hr.
Cells were then rinsed, placed on ice for 10 min, and incubated with 10–25 µg/ml Alexa 594-conjugated Transferrin or cholera toxin subunit B diluted in supplemented DMEM for 15–20 min at 37˚C prior to fixation and imaging.
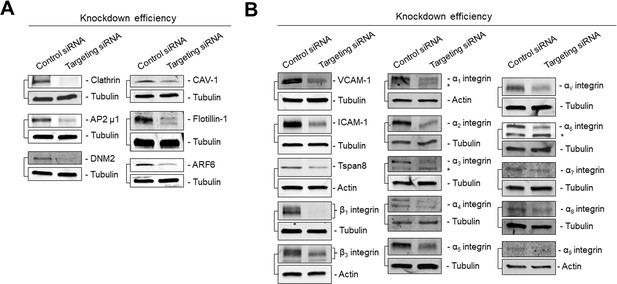
Immunoblots showing siRNA-mediated knockdown efficiencies corresponding to data of Figure 1.
(A) Immunoblots showing knockdown efficiency of siRNA transfections related to Figure 1B. (B) Immunoblots showing knockdown efficiency of siRNA transfections related to Figure 1D.
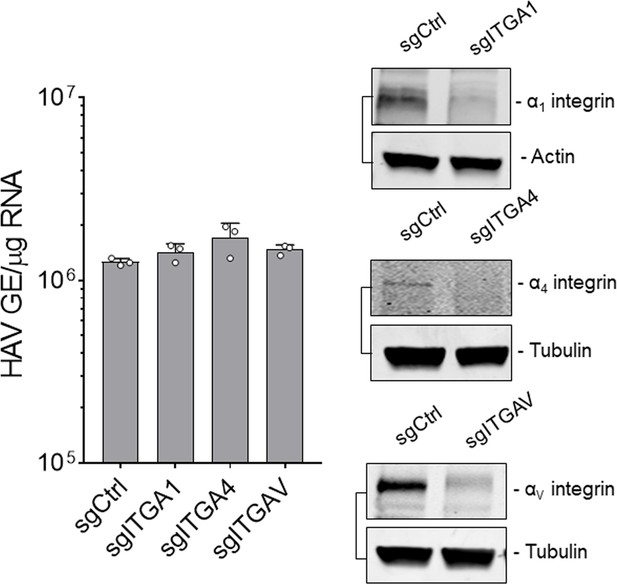
Levels of HAV RNA at 24 hpi in H1-HeLa cells engineered by CRISPR/Cas9 to be knocked-out for the indicated integrins inoculated with naked HAV (mean ± SD).
Immunoblots of whole-cell lysates harvested from the indicated cell lines.
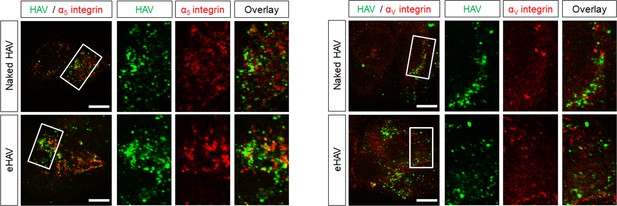
Huh-7.5 cells were inoculated with HAV or eHAV and subjected to immunostaining one hpi using antibodies against HAV capsid (K24F2) and α5 or αV integrins.
Scale bar, 10 µm.
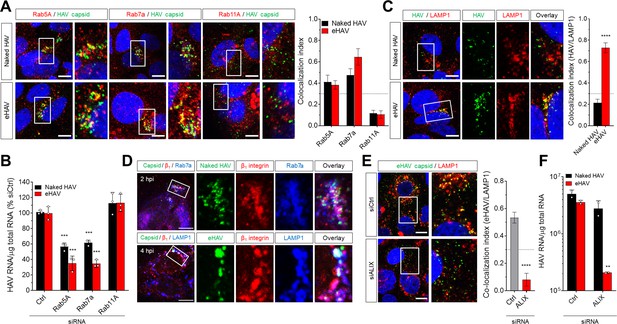
Distinct endocytic sorting of naked and quasi-enveloped HAV.
(A) Confocal micrographs of Huh-7.5 cells immunolabeled with anti-HAV capsid (K24F2) and anti-Rab5A, Rab7a, or Rab11A at two hpi. Scale bar, 10 µm. (B) Effect of siRNA-mediated depletion of Rab GTPases on HAV and eHAV entry (mean ± SD, n = 3 independent experiments). See Figure 2—figure supplement 1 for knockdown efficiencies. (C) Confocal micrographs of Huh-7.5 cells immunolabeled with anti-HAV capsid (K24F2) and anti-LAMP1 at six hpi. Scale bar, 10 µm. (D) Confocal micrographs of Huh-7.5 cells adsorbed with naked HAV or eHAV and immunolabeled with antibodies against HAV capsid (K24F2), β1 integrin, and either Rab7 or LAMP1. Scale bar, 10 µm. (E) Confocal micrographs of Huh-7.5 cells previously transfected with a control or ALIX-specific siRNAs and immunolabeled with anti-HAV capsid (K24F2) and anti-LAMP1 at 12 hpi with eHAV. Scale bar, 10 µm. See Figure 2—figure supplement 5 for knockdown efficiencies. (F) Effect of ALIX depletion by siRNA on eHAV and HAV entry and replication at 22 hpi (mean ± SD, n = 2 biological replicates for a representative experiment). For numeric data plotted in graphs associated with this figure, see Figure 2—source data 1.
-
Figure 2—source data 1
Source data corresponding to Figure 2.
- https://doi.org/10.7554/eLife.43983.015
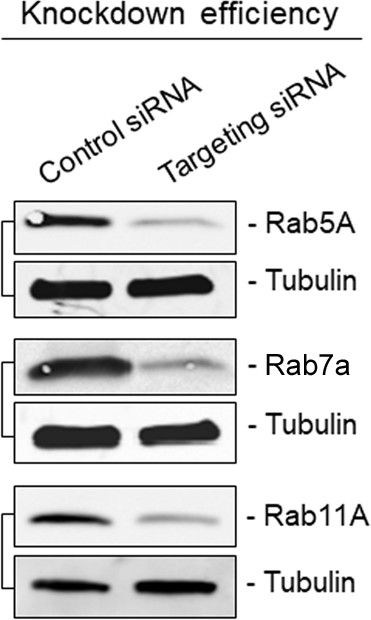
Immunoblots showing knockdown efficiency of siRNA transfections related to Figure 2B.
https://doi.org/10.7554/eLife.43983.010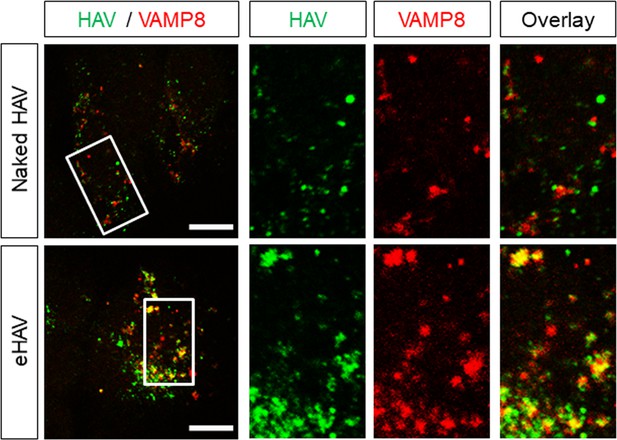
Huh-7.5 cells were inoculated with gradient-purified HAV or eHAV, fixed 6 hr post-adsorption, and subjected to immunostaining using antibodies against HAV capsid (K24F2) and the lysosomal protein VAMP8.
Scale bar, 10 µm.
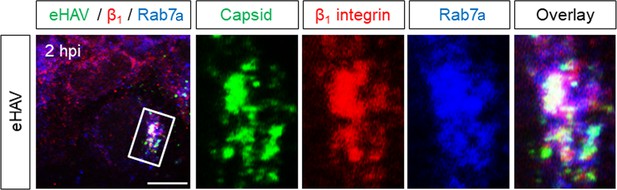
Huh-7.5 cells were inoculated with eHAV, fixed two hpi, and subjected to immunostaining using antibodies against HAV capsid (K24F2), β1 integrin, and Rab7.
Scale bar, 10 µm.
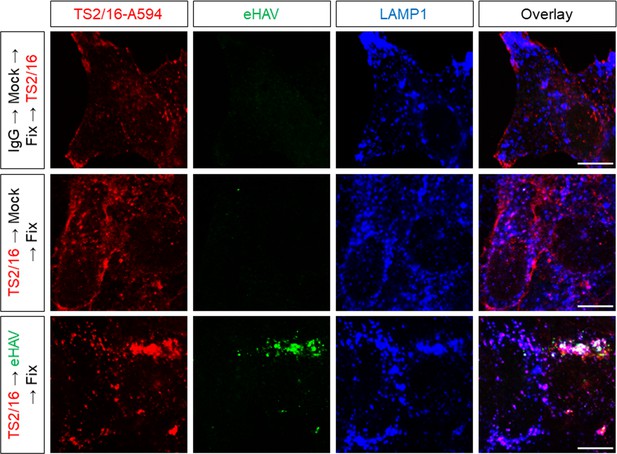
Huh-7.5 cells were pre-treated with the Alexa594-conjugated activating anti-β1 integrin mAb -TS2/16 or IgG control for 20 min on ice to label cell surface β1 integrin prior to virus adsorption.
Inoculum was removed after 1 hr and cells were fixed and immunostained 5 hr later with anti-HAV (JC polyclonal human sera) and anti-LAMP1. Scale bar, 10 µm.
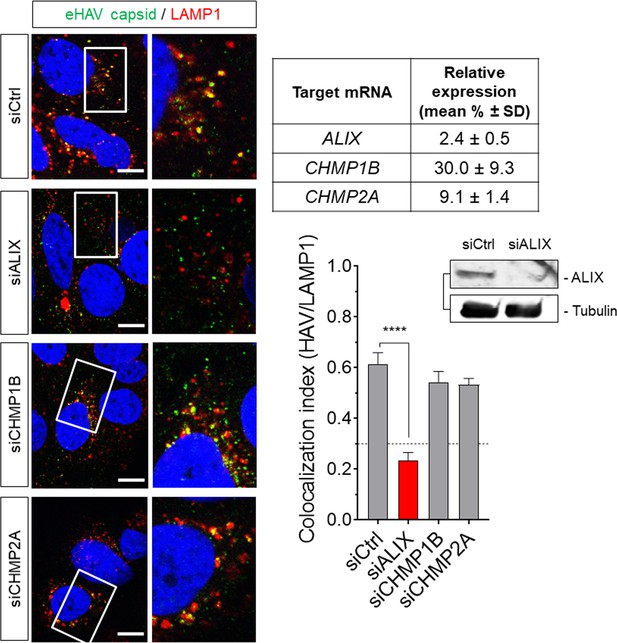
Huh-7.5 cells were transfected with the indicated siRNAs for 72 hr prior to adsorption with eHAV.
Cells were immunostained at eight hpi using anti-HAV capsid (K24F2) and anti-LAMP1. Scale bar, 10 µm. Knockdown efficiency was determined by immunoblotting or by RT-qPCR (table) using gene-specific primers to obtain calculate expression levels relative to control siRNA-transfected cells.
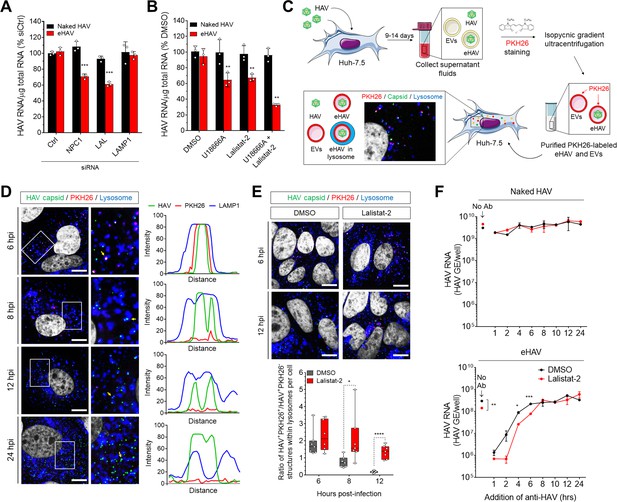
Degradation of the eHAV quasi-envelope occurs within the lysosome.
(A) Effect of siRNA-mediated depletion of lysosomal-associated proteins on HAV and eHAV entry (mean ± SD, n = 3 independent experiments). See Figure 3—figure supplement 1 for knockdown efficiencies. (B) Effect of inhibition of NPC1 or LAL by U18666A or Lalistat-2, respectively, on HAV and eHAV entry (mean ± SD, n = 3 independent experiments). (C) Strategy for membrane labeling of exosomes and eHAV with PKH26 dye. (D) Confocal micrographs of Huh-7.5 cells inoculated with PKH26-labeled eHAV and immunostained with anti-HAV capsid (K24F2) and anti-LAMP1. Histograms depict pixel intensities for the structures indicated with a yellow arrow. Scale bar, 10 µm. (E) Confocal micrographs of Huh-7.5 cells inoculated with PKH26-labeled eHAV in presence of Lalistat-2 and immunostained with anti-HAV capsid (K24F2) and anti-LAMP1. Graph displays the ratio of structures corresponding to PKH26-associated HAV capsid (green + red signal) over HAV capsids devoid of PKH26 fluorescence (green alone) within lysosomes per cell. Scale bar, 10 µm (F) Effect of Lalistat-2 on post-endocytic antibody-mediated neutralization of eHAV. Neutralizing anti-HAV (JC antibody) was added at the indicated intervals after removal of the inoculum and intracellular viral RNA was quantified at 48 hr (mean ±SD, n = 2 biological replicates for a representative experiment). For numeric data plotted in graphs associated with this figure, see Figure 3—source data 1.
-
Figure 3—source data 1
Source data corresponding to Figure 3.
- https://doi.org/10.7554/eLife.43983.019
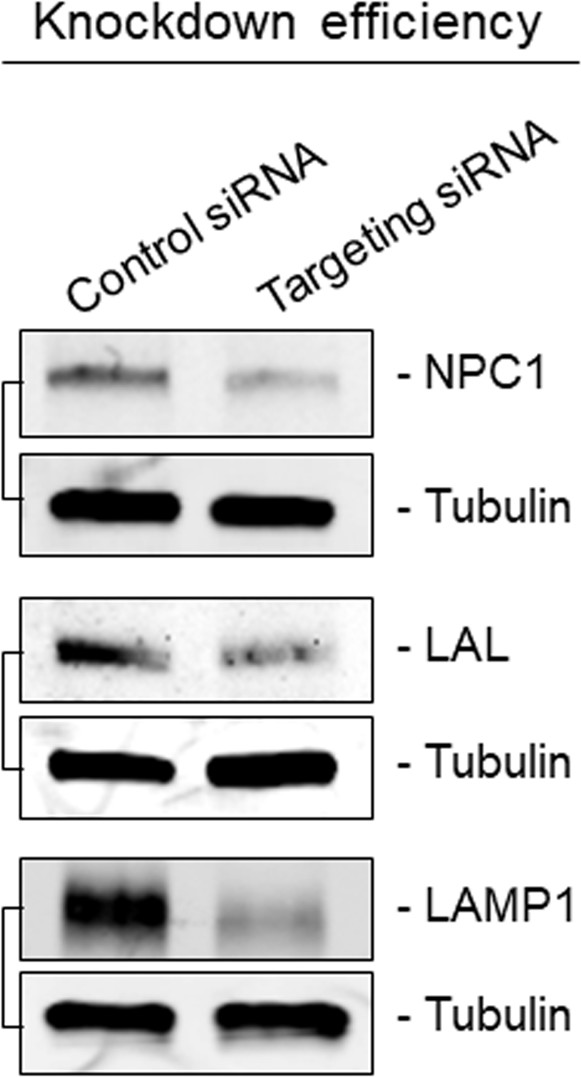
Immunoblots showing knockdown efficiency of siRNA transfections related to Figure 3A.
https://doi.org/10.7554/eLife.43983.017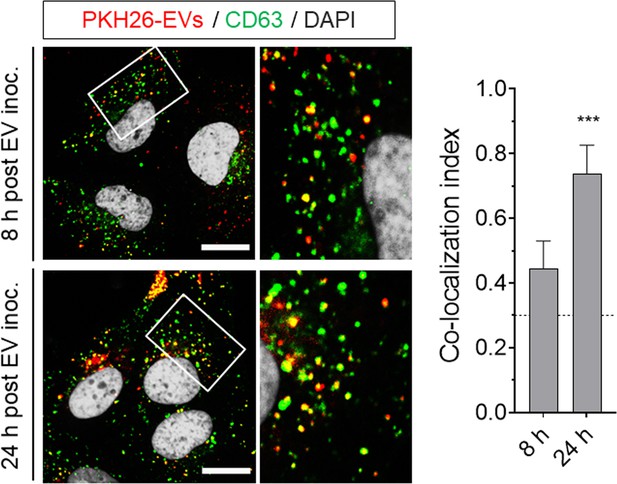
Huh-7.5 cells were adsorbed with PKH26-labeled gradient-purified extracellular vesicles (EVs, fractions 9 to 11 in iodixanol gradient) obtained from supernatant fluids of uninfected Huh-7.5 cells, inocula was removed after 1 hr, and cells were fixed and immunostained with anti-CD63 at 8 or 24 hr post-inoculation.
Scale bar, 10 µm.
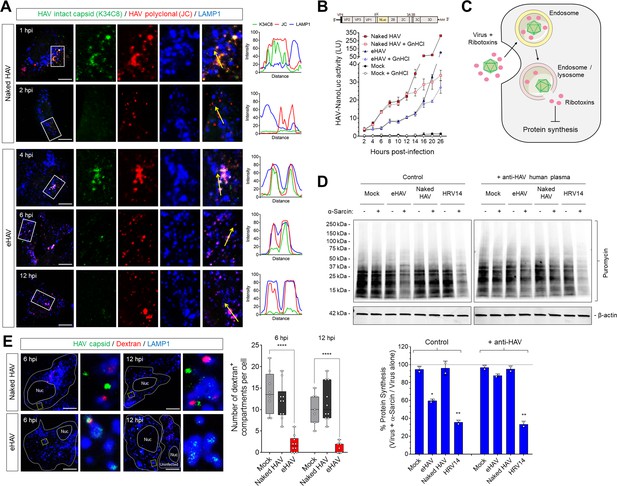
Different uncoating mechanisms for naked and quasi-enveloped HAV.
(A) Confocal micrographs of Huh-7.5 cells adsorbed with HAV or eHAV in presence of cycloheximide immunostained with an antibody that recognizes intact HAV capsids (K34C8), a polyclonal human anti-HAV that recognizes denatured P1 proteins (JC), and LAMP1. Histograms depict pixel intensities for the structures indicated with a yellow arrow. Scale bar, 10 µm. (B) Kinetics of HAV RNA translation measured by nanoluciferase production by HAV-NLuc in H1-HeLa cells pre-treated or not with GnHCl to inhibit viral replication. (C) Schematic of the endolysosomal permeabilization assay using endocytosed ribotoxins. (D) Virus-induced endolysosomal membrane damage at six hpi. H1-HeLa cells were pulsed for 20 min with puromycin prior to cell lysis, and protein synthesis was analyzed by immunoblotting using an anti-puromycin antibody. Alternatively, viruses were incubated overnight at 4˚C with human ‘JC’ plasma containing neutralizing HAV antibodies prior to adsorption and adsorbed as a mix. Band intensities were normalized to actin and protein synthesis is expressed as band intensity in cells infected in presence of α-sarcin relative to cells infected with virus alone (mean ± SD from two independent cultures of a representative of four independent experiments). (E) Micrographs of Huh-7.5 cells with lysosomes pre-loaded with fluorescently-conjugated dextran (10 kDa) prior to adsorption with HAV or eHAV and immunostained using antibodies against HAV capsid (K24F2) and LAMP1. Graphs display the number of dextran +compartments per cell (n = 7–10 cells per condition for a representative of three independent experiments). Scale bar, 10 µm. For numeric data plotted in graphs associated with this figure, see Figure 4—source data 1.
-
Figure 4—source data 1
Source data corresponding to Figure 4.
- https://doi.org/10.7554/eLife.43983.024
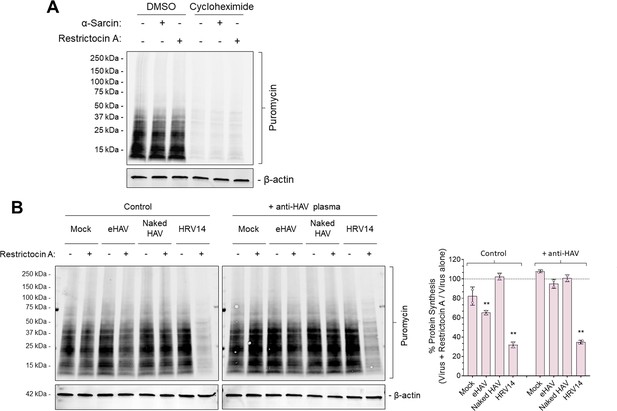
Validation of puromycin incorporation into nascent proteins and Restrictocin A-induced inhibition of protein synthesis during viral entry.
(A) H1-HeLa cells were incubated with 100 µg/ml α-sarcin or 50 µg/ml Restrictocin A for 6 hr at 37˚C, followed by treatment with cycloheximide for 30 min prior to a 20 min pulse with 20 µg/ml puromycin. Protein lysates were harvested, resolved by SDS-PAGE and subjected to immunoblotting using antibodies against puromycin and actin. (B) H1-HeLa cells were adsorbed for 1 hr with gradient-purified eHAV or naked HAV (~1000 GEs/cell) at 37˚C or with HRV14 (10 PFU/cell) at 33˚C in presence or absence of 50 µg/ml Restrictocin A. The inoculum was removed, cells were rinsed with PBS, and incubated for another 5 hr. Cells were then pulsed with puromycin for 20 min and protein lysates were harvested, resolved by SDS-PAGE, and subjected to immublobotting using antibodies against puromycin and actin. Band intensities were quantified, normalized to loading control, and presented as a percentage of virus plus α-sarcin relative to virus alone. Alternatively, viruses were incubated overnight at 4˚C with human ‘JC’ plasma containing neutralizing HAV antibodies and inoculated as such.
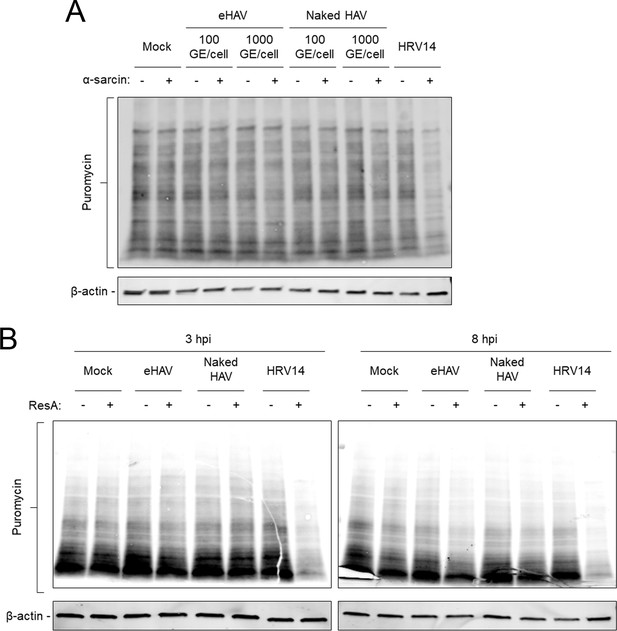
Kinetics of virus-induced endosomal membrane damage.
(A) Cells were adsorbed for 1 hr with gradient-purified eHAV or naked HAV (~100 or~1000 GEs/cell) at 37˚C or with HRV14 (10 PFU/cell) at 33˚C in presence or absence of α-sarcin followed by a puromycin pulse for 20 min. Protein lysates were harvested and resolved by SDS-PAGE and subjected to immunoblotting using antibodies against puromycin and actin. (B) H1-HeLa cells were adsorbed for 1 hr with gradient-purified eHAV or naked HAV (~1000 GEs/cell) at 37˚C or with HRV14 (10 PFU/cell) at 33˚C in presence or absence of Restrictocin A. The inoculum was removed, cells were rinsed with PBS, and incubated for an additional 2 or 7 hr. Cells were then pulsed with 20 µg/ml puromycin for 20 min, protein lysates were harvested, resolved by SDS-PAGE, and subjected to immunoblotting using antibodies against puromycin and actin.
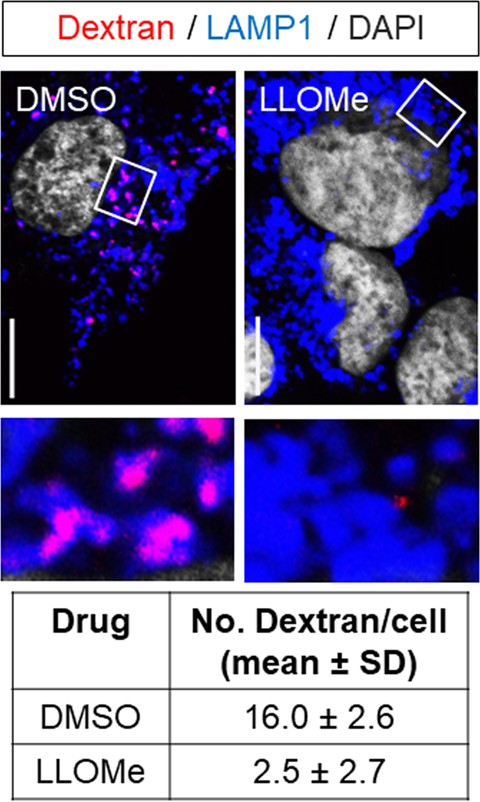
Lysosomes of Huh-7.5 cells were pre-loaded with fluorescently-conjugated dextran (10 kDa) overnight and cells were incubated with DMSO control or LLOMe for 2 hr prior to fixation and immunostaining with anti-LAMP1.
Scale bar, 10 µm.
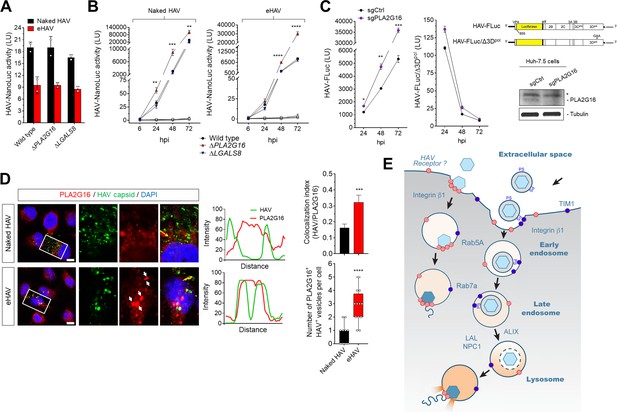
Dispensability of PLA2G16 during HAV entry.
(A) Translation of HAV RNA during viral entry (~2000 GEs/cell, 12 hpi) measured by HAV-NLuc activity in H1-HeLa cells knocked-out for PLA2G16 or LGALS8 (mean ± SD, n = 2 biological replicates for a representative experiment). (B) Replication kinetics of naked or quasi-enveloped HAV/NLuc in H1-HeLa cells (mean ± SD, n = 3 biological replicates for a representative experiment). (C) Graphs showing luciferase activity expressed by transfected HAV-FLuc subgenomic replicon RNA (left) or a replication-incompetent mutant (right) in sgControl or sgPLA2G16 Huh-7.5 cells (mean ± SD, n = 2 biological replicates for a representative experiment). At the lower right is an immunoplot of PLA2G16 in these cells (*non-specific band). (D) Micrographs of H1-HeLa cells immunolabeled with anti-HAV capsid (K24F2) and anti-PLA2G16 at six hpi with naked HAV or eHAV. White arrows indicate sites of presumed membrane damage where PLA2G16 has accumulated. Scale bar, 10 µm. Histograms depict pixel intensities across the drawn arrow on the inset overlay. Graphs show the co-localization (Mander’s coefficients) (n = 8 cells per condition) between HAV and PLA2G16 or the number of vesicles positive for both HAV capsid and PLA2G16. (E) Current model for cellular entry of naked and quasi-enveloped HAV virions. PS: phosphatidylserine. For numeric data plotted in graphs associated with this figure, see Figure 5—source data 1.
-
Figure 5—source data 1
Source data corresponding to Figure 5.
- https://doi.org/10.7554/eLife.43983.028
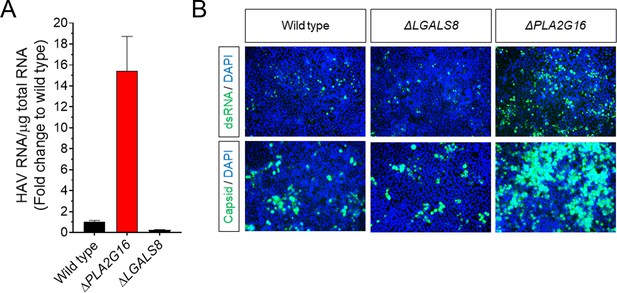
Restriction of HAV replication by PLA2G16 in H1-HeLa cells.
(A) H1-HeLa cells were inoculated with naked HAV (~1 GE/cell) and intracellular RNA was harvested four days post-infection (mean fold HAV RNA ± SD). (B) H1-HeLa cells were inoculated with naked HAV (~1 GE/cell), fixed 5 days post-infection, and immunostained with antibodies against dsRNA (J2 clone mAb) or HAV capsid (K24F2).
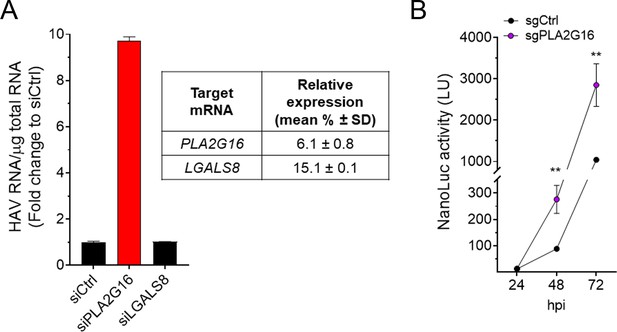
Restriction of HAV replication by PLA2G16 in Huh-7.5 cells.
(A) Huh-7.5 cells were transfected with the indicated siRNAs for 72 hr, infected with naked HAV (~10 GEs/cell) and intracellular RNA was harvested 72 hpi (mean fold HAV RNA ± SD). (B) Huh-7.5 cells were infected with cell-free HAV-NLuc and nanoluciferase production was obtained at the indicated hours post-infection (mean ± SD).
Tables
| Reagent type (species) | Designation | Source/reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | Huh-7.5 | PMID: 12584342 | RRID: CVCL_7927 | Gift from Charles Rice |
| Cell line (human) | HeLa ΔPLA2G16 | PMID: 28077878 | Gift from Thijn Brummelkamp | |
| Cell line (human) | HeLa ΔGALS8 | PMID: 28077878 | Gift from Thijn Brummelkamp | |
| Biological sample (virus) | HM175/18 f hepatitis A virus (HAV) | PMID: 1705995 | GenBank KP879216.1 | |
| Biological sample (virus) | HM175/p16 hepatitis A virus (HAV) | PMID: 2833008 | GenBank KP879217.1 | |
| Biological sample (virus) | Human rhinovirus 14 (HRV14) | PMID: 8383233 | Recovered from pWR3.26; gift from Roland Rueckert | |
| Antibody (rabbit) | anti-clathrin heavy chain | Abcam | ab21679 | (WB) 1:2000, (IF) 1:1000 |
| Antibody (rabbit) | anti-AP2M1 | GeneTex | GTX113332 | (WB) 1:1000 |
| Antibody (rabbit) | anti-DNM2 | GeneTex | GTX113171 | (WB) 1:1000 |
| Antibody (rabbit) | anti-CAV1 | Abcam | ab2910 | (WB) 1:2000, (IF) 1:500 |
| Antibody (rabbit) | anti-FLOT1 | GeneTex | GTX104769 | (WB) 1:1000 |
| Antibody (rabbit) | anti-ARF6 | GeneTex | GTX112872 | (WB) 1:1000 |
| Antibody (rabbit) | anti-Rab5A | Abcam | ab18211 | (WB) 1:1000 |
| Antibody (rabbit, mAb) | anti-Rab7a | Cell Signaling | 9367 | (WB) 1:1000, (IF) 1:100 |
| Antibody (rabbit, mAb) | anti-Rab11A | Cell Signaling | 5589 | (WB) 1:1000, (IF) 1:100 |
| Antibody (rabbit, mAb) | anti-NPC1 | Abcam | ab134113 | (WB) 1:2000 |
| Antibody (rabbit) | anti-LAL | GeneTex | GTX101169 | (WB) 1:1000 |
| Antibody (mouse, mAb) | anti-ALIX | Santa Cruz | sc-53540 | (WB) 1:500 |
| Antibody (mouse, mAb) | anti-puromycin | Millipore | MABE343 | (WB) 1:10000 |
| Antibody (rabbit) | anti-PLA2G16 | Cayman Chemical | 10337 | (WB) 1:200 |
| Antibody (goat) | anti-Galectin-8 | R and D Systems | AF1305 | (WB) 1:500 |
| Antibody (mouse, mAb) | anti-tubulin | Sigma | T6199 | (WB) 1:20000 |
| Antibody (rabbit) | and anti-actin | Sigma | A2066 | (WB) 1:5000 |
| Antibody (sheep) | anti-VCAM-1 | R and D Systems | AF809 | (WB) 1:250 |
| Antibody (sheep) | anti-ICAM-1 | R and D Systems | AF720 | (WB) 1:250 |
| Antibody (rat, mAb) | anti-Tspan8 | R and D Systems | MAB4734 | (WB) 1:150 |
| Antibody (rabbit, mAb) | anti-integrin β1 | Cell Signaling | 9699 | (WB) 1:1000 |
| Antibody (rabbit, mAb) | anti-integrin β3 | Cell Signaling | 13166 | (WB) 1:500 |
| Antibody (rabbit, mAb) | anti-integrin α1 | R and D Systems | MAB5676 | (WB) 1:250 |
| Antibody (rabbit, mAb) | anti-integrin α2 | Abcam | ab133557 | (WB) 1:500 |
| Antibody (rabbit) | anti-integrin α3 | Millipore | AB1920 | (WB) 1:250 |
| Antibody (rabbit, mAb) | anti-integrin α4 | Cell Signaling | 8440 | (WB) 1:250 |
| Antibody (rabbit) | anti-integrin α5 | Cell Signaling | 4705 | (WB) 1:700 |
| Antibody (rabbit) | anti-integrin αV | Cell Signaling | 4711 | (WB) 1:700 |
| Antibody (rabbit) | anti-integrin α6 | GeneTex | GTX100565 | (WB) 1:500 |
| Antibody (rabbit) | anti-integrin α7 | Thermo Fisher Sci | PA5-37435 | (WB) 1:250 |
| Antibody (rabbit) | anti-integrin α8 | Novus Biologicals | NBP1-59940 | (WB) 1:250 |
| Antibody (mouse, mAb) | anti-integrin α9 | R and D Systems | MAB4574 | (WB) 1:250 |
| Antibody (mouse, mAb) | anti-integrin β1 | Abcam | ab30394 | (IF) 1:100 |
| Antibody (rabbit, mAb) | anti-integrin α5 | Abcam | ab150361 | (IF) 1:250 |
| Antibody (rabbit, mAb) | anti-integrin αV | Abcam | ab179475 | (IF) 1:500 |
| Antibody (rabbit, mAb) | anti-Rab11 | Cell Signaling | 5589 | (IF) 1:100 |
| Antibody (rabbit, mAb) | anti-LAMP1 | Cell Signaling | 9091 | (IF) 1:200 |
| Antibody (rabbit, mAb) | anti-VAMP8 | Abcam | ab76021 | (IF) 1:250 |
| Antibody (mouse, mAb) | anti-CD63 | BD Biosciences | #556019 | (IF) 1:50 |
| Antibody (rabbit) | anti-PLA2G16 | Sigma | H8290 | (IF) 1:50 |
| Antibody (mouse, mAb) | anti-dsRNA | Scicons | J2 | (IF) 1:1000 |
| Antibody (human) | JC human anti-HAV | PMID: 23542590 | JC | (IF) 1:600 |
| Antibody (mouse, mAb) | K24F2 anti-HAV capsid | PMID: 6315771 | K24F2 | (IF) 1:100 |
| Antibody (mouse, mAb) | K34C8 anti-HAV capsid | MacGregor et al. | K34C8 | (IF) 1:300 |
| Antibody (mouse, mAb) | anti-integrin β1 K-20 | Santa Cruz | sc-18887 | 10 µg ml−1 |
| Antibody (mouse, mAb) | anti-integrin β1 TS2/16 | Santa Cruz | sc-53711 | 10 µg ml−1 |
| Antibody (mouse, mAb) | anti-integrin β1 8E3 | Millipore | MABT199 | 10 µg ml−1 |
| Antibody (mouse, mAb) | anti-integrin β1 HUTS-2 | Millipore | MAB2079Z | 10 µg ml−1 |
| Chemical compound | dynasore | Millipore | #324410 | 80 µM |
| Chemical compound | chlorpromazine | Sigma-Aldrich | #C8138 | 10 µg ml−1 |
| Chemical compound | filipin | Sigma-Aldrich | #F9765 | 1 µg ml−1 |
| Chemical compound | cytochalasin D | Sigma-Aldrich | #C2618 | 20 µM |
| Chemical compound | latrunculin A | Sigma-Aldrich | #428026 | 1 µM |
| Chemical compound | 5-(N-Ethyl-N- isopropyl)amiloride) | Sigma-Aldrich | #A3085 | 1 µM |
| Chemical compound | wortmannin | Sigma-Aldrich | #W1628 | 1 µM |
| Chemical compound | NSC23766 | Sigma-Aldrich | #SML0952 | 300 µM |
| Chemical compound | dynarrestin | PMID: 29396292 | 25 µM, gift from Jared Sterneckert | |
| Chemical compound | heparin | Sigma-Aldrich | #H3149 | 10 µg ml−1 |
| Chemical compound | Lalistat-2 | PMID: 20557099 | 200 µM, gift from Paul Helquist | |
| Chemical compound | U18666A | Sigma-Aldrich | #U3633 | 2 µg ml−1 |
| Chemical compound | guanidine hydrochloride | Sigma | #G3272 | 5 mM |
| Chemical compound | cycloheximide | Sigma | #C7698 | 25 µg ml−1 |
| Chemical compound | puromycin | InvivoGen | CAS 58-58-2 | 5–20 µg ml−1 |
| Chemical compound | α-sarcin | Santa Cruz | #sc-204427 | 100 µg ml−1 |
| Chemical compound | Restrictocin A | Sigma-Aldrich | #R0389 | 50 µg ml−1 |
| Chemical compound | L-Leucyl-L-Leucine- methyl ester | Cayman Chemical | #16008 | 2 mM |
Additional files
-
Supplementary file 1
Nucleotide sequences of sgRNAs, gene-specific RT-PCR primers and siRNAs.
- https://doi.org/10.7554/eLife.43983.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43983.030





