Distinct origins and molecular mechanisms contribute to lymphatic formation during cardiac growth and regeneration
Figures
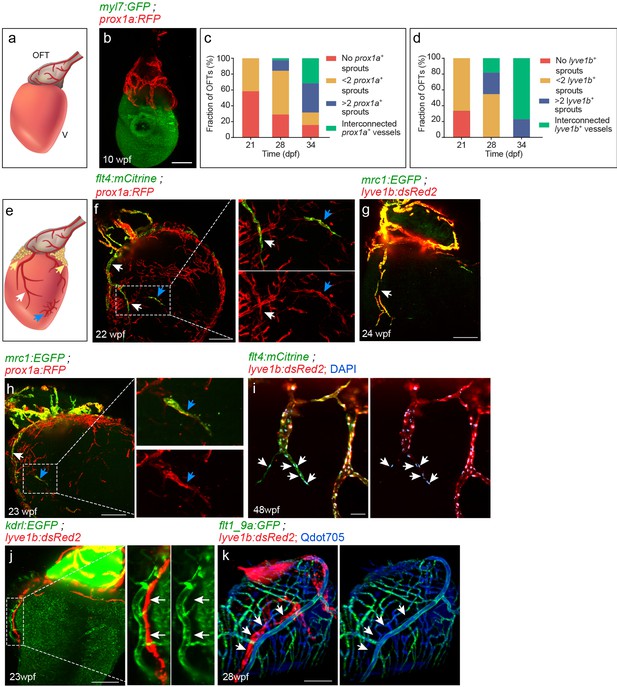
Lymphatic vessel heterogeneity in the zebrafish heart.
(a) Diagram of ~7 wpf zebrafish heart depicting the outflow tract (OFT), ventricle (V) and lymphatics (red). (b) At 10 wpf (fish size 16–24 mm) prox1a-labeled collecting lymphatics are clearly detected in the OFT, but absent on the ventricular surface (n = 9). (c,d) Quantification of 21–34 dpf (fish size 7–14 mm) OFT lymphatic development in prox1a (c) (n21dpf=48, n28dpf=38, n34dpf=19) andlyve1b (d) (n21dpf=12, n28dpf=11, n34dpf=13) transgenic zebrafish. (e) Diagram of adult zebrafish heart depicting ventricular lymphatics (white arrows), fat-associated lymphatics (yellow arrows) and isolated lymphatic clusters (blue arrows). OFT and ventricular lymphatics as well as isolated lymphatic clusters in 22–24 wpf (fish size 25–30 mm) are labeled by the flt4 (f), lyve1b (g), prox1a (f,h) and mrc1a (g,h) (nf = 4, ng = 5, nh = 5) transgenic reporters. (i) 1–5 cells at the tip of ventricular lymphatics are labeled primarily by the flt4 transgene (arrows). Nuclei are labeled by DAPI (blue) (48wpf, fish size 25–30 mm, n = 5). (j) Ventricular lymphatics are not labelled by the blood vessel/endocardial- marker Tg(kdrl:nls-mCherry) (23wpf, fish size 25–30 mm, n = 5). (k) Angiogram of 28 wpf (fish size 28 mm) Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed2) heart. Cardiac lymphatics (arrows) are not labeled following intravascular injection of Qdot705 (blue) (n = 6). Scale bars are 200 µm in b, f-h, j, k; 50 µm in i. Posterior view in b, anterior view in f-k.
-
Figure 1—source data 1
Development of OFT lymphatics.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig1-data1-v3.xlsx
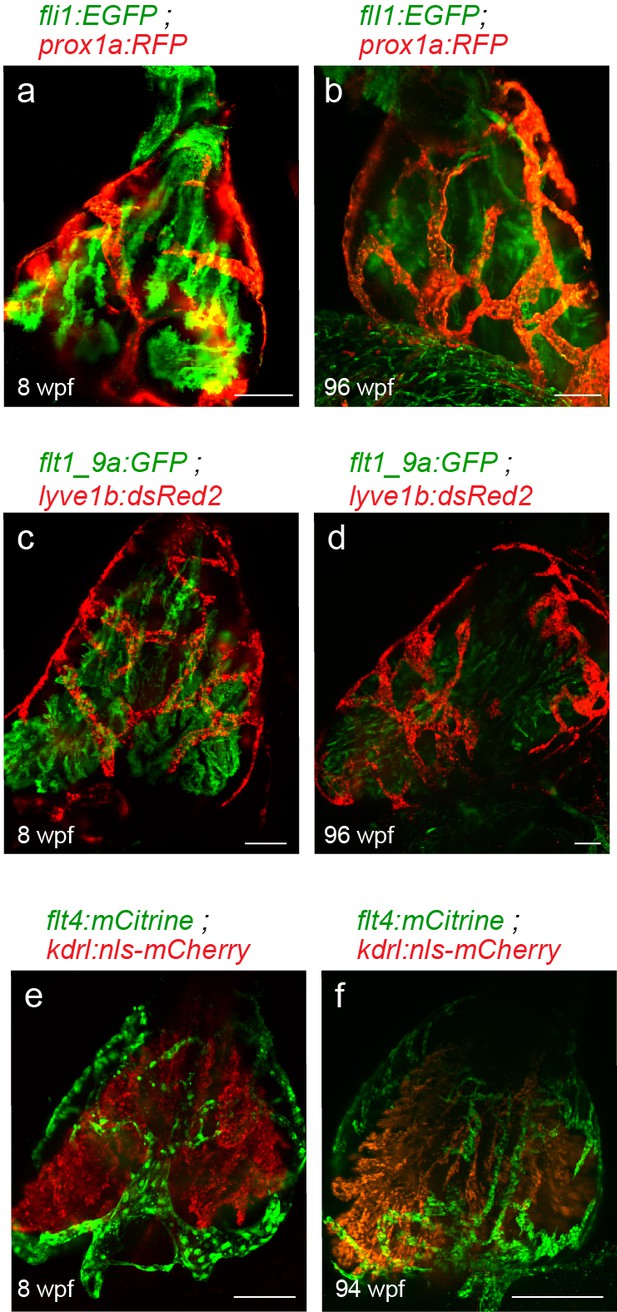
OFT lymphatics are fully established by eight wpf.
OFT of 8 wpf (fish size 14–24 mm) (a,c,e) and 96 wpf (fish size 25–32 mm) (b,d,f) Tg(fli1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) (a,b), Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed2) (c,d) and Tg(kdrl:nls-mCherry);Tg(flt4BAC:mCitrine) (e,f) showing well-established lymphatic vessels, which remain stable through adulthood. Scale bars are 100 µm. Posterior view.
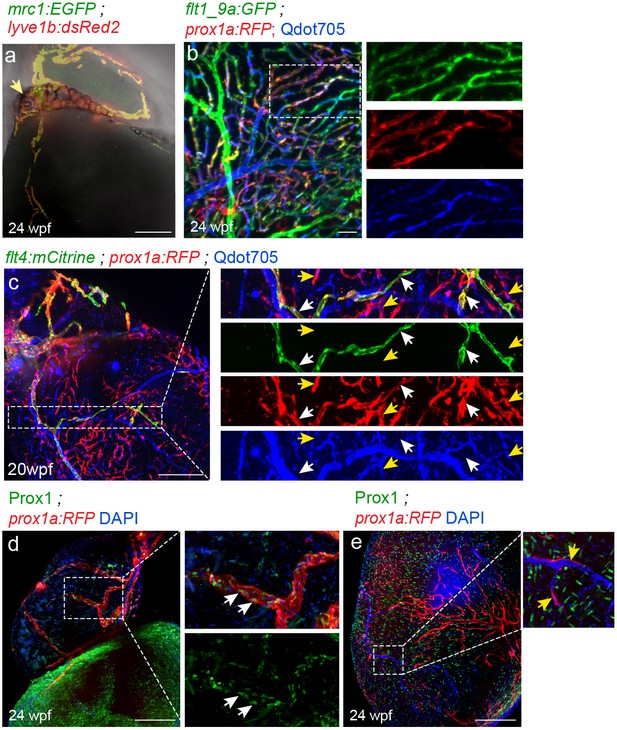
Prox1a transgene labels cardiac lymphatics as well as a subset of coronary vessels.
(a) Lymphatics associated with cardiac fat tissue (yellow arrow), in 24 wpf (fish size 29 mm) Tg(mrc1a:EGFP);Tg(lyve1b:dsRed2) animals presented in Figure 1g. Light sheet image super imposed on a bright-field image (n = 3). (b) prox1a transgene is detected in blood vessels labeled by the flt1_9a arterial marker and highlighted following intravascular injection of Qdot705 (blue) (n = 4) (insets). (c) intravascularly injected Qdot705 is detected in blood capillaries labeled by prox1a only (yellow arrows), but not in double-positive flt4;prox1a lymphatic vessels (white arrows). (d,e) Prox1 immunostaining (green) confirms Prox1 expression in OFT lymphatics (d, inset, arrows) but not in prox1a transgene labeled (red) blood vessels (e, inset, yellow arrows). Erythrocyte nuclei inside prox1a-positive blood vessels are labeled by DAPI (blue). Prox1 also labels cardiomyocytes (n = 3). Scale bars are 200 µm. Anterior view in a,b,c,e, lateral view in d.
Individual Z-stacks through heart shown in Figure 1h confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(mrc1a:EGFP) is in green.
Individual Z-stacks through heart shown in Figure 1f confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt4BAC:mCitrine) is in green.
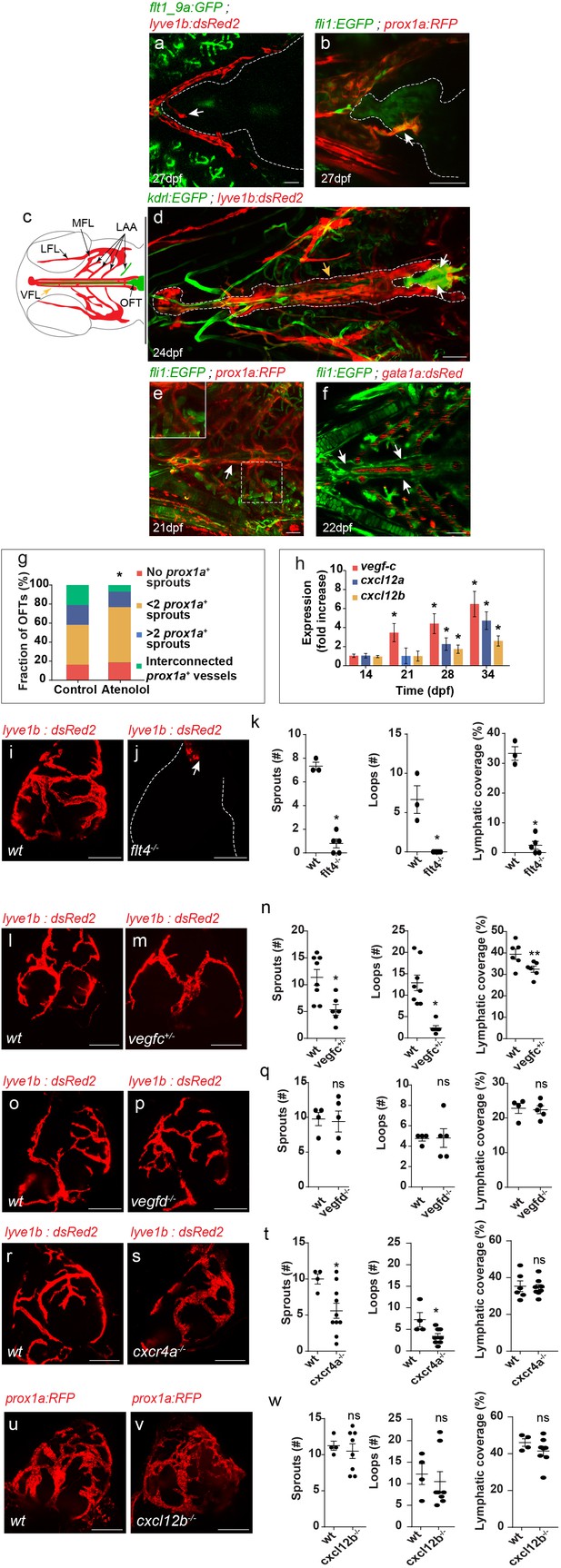
Establishment of OFT lymphatics.
(a–b) Blood vessels are not detected in the OFT (outlined) of 21–28 dpf (fish size 5–7 mm) (a) Tg(flt1_9a_cFos:GFP); Tg(lyve1b:dsRed2) hearts (n = 6) or (b) Tg(fli1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) (n = 4) fish, prior to OFT lymphatic sprouting (white arrows). (c) Diagram depicting a ventral view of a zebrafish larval head, indicating the approximate region imaged in (d-f). Facial lymphatics are colored in red (adapted from Okuda et al., 2012), yellow arrow points to the VFL. (d) OFT lymphatic sprouts (white arrows) arising from the VFL (outlined, yellow arrow) are detected at 24 dpf (Fish size 5–7 mm) in Tg(kdrl:EGFP);Tg(lyve1b:dsRed2);casper larvae (n = 5). (e) The VFL (arrow) connects to the LAA (inset). (f) No blood flow is detected in the VFL (arrows) of 22 dpf (fish size 5—7 mm) Tg(fli:EGFP);Tg(gata1a:dsRed2) larvae (n = 10). (g) Quantification of OFT lymphatics in 35 dpf (fish size 9–13 mm) prox1a transgenic zebrafish treated with 100 µM Atenolol (ncontrol = 43, nAtenolol = 43, *p<0.001). (h) mRNA levels of vegfc, cxcl12a and cxcl12b (nindependent experiments=5, *p<0.01) in the OFTs of 14–34 dpf larvae. (i–k) OFT of 19 wpf (22–25 mm) wt sibling (i) and flt4-/- (j) in the background of lyve1b demonstrating severe lymphatic defects in flt4 -/- hearts, quantified in (k) (nwt = 4, nflt4 -/-=5, *p<0.001). (l–n) OFT of 15 wpf (20–21 mm) wt sibling (l) and vegfc +/- (m) in the background of lyve1b showing malformed lymphatics in vegfc +/-, quantified in (n) (nwt = 8, nvegfc+/-=6, *p<0.01, **p<0.05). (o–q) OFT of 12 wpf (19–21 mm) age-matched wt control (o) and vegfd -/- (p) in the background of lyve1b showing normal OFT lymphatics in vegfd -/- hearts, quantified in (q) (nwt = 4, nvegfd-/-=5) (r–t) OFT lymphatics of 9.5 wpf (20–23 mm) wt sibling (r) and cxcr4a-/- (s) in the background of lyve1b showing mild defects in cxcr4a-/- OFT lymphatics, quantified in (t) (nwt = 4, ncxcr4a-/-=10, *p<0.01). (u–w) OFT lymphatics of 20 wpf (19–24 mm) wt sibling (u) and cxcl12b-/- (v) in the background of prox1a showing normal OFT lymphatics in cxcl12b-/- hearts, quantified in (w) (nwt = 4, ncxcl12b -/-=8). VFL, ventral facial lymphatics; LFL, lateral facial lymphatic; LAA, lymphatic branchial arches; MFL, medial facial lymphatic. Scale bars are 50 µm in a-f, 200 µm in i-v. Error bars, mean ± S.E.M.
-
Figure 2—source data 1
Quantification of OFT lymphatics following Atenolol treatment.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig2-data1-v3.xlsx
-
Figure 2—source data 2
mRNA levels of pro-lymphangiogenic cues in the OFTs of 14–34 dpf larvae.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig2-data2-v3.xlsx
-
Figure 2—source data 3
Development of OFT lymphatics in flt4-/-, vegfc+/-, vegfd -/-, cxcr4a -/- and cxcl12b -/- hearts.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig2-data3-v3.xlsx
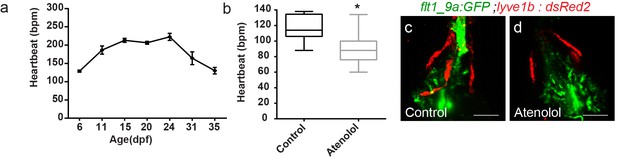
Hemodynamic changes during larva to juvenile transition modulate OFT lymphatic development.
(a) Heart rate peaks at ~21 dpf in zebrafish larvae. (b) Heartbeat of 35 dpf fish is reduced following treatment with 100 µM atenolol. (c,d) OFT lymphatics development is delayed in 35 dpf atenolol (d) treated fish as compared to sibling control (c) (ncontrol = 14, nAtenolol = 15, *p<0.001). Scale bars are 50 µm.
-
Figure 2—figure supplement 1—source data 1
Quantification of larvae heartbeat during development and following Atenolol treatment.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig2-figsupp1-data1-v3.xlsx
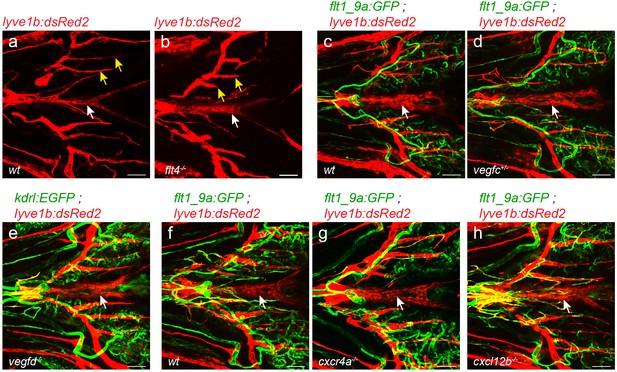
VFL develops normally in lymphatic-related mutants.
(a–h) ventral view of 22dpf (fish size 5–7 mm) fish. (a,b) while medial facial lymphatics are shorter in Tg(lyve1b:dsRed2);flt4-/- as compared to wt siblings (yellow arrows), VFL forms normally (white arrow). Normal VFL is also detected in vegfc+/- heterozygous (c,d), vegfd-/- (e,f) cxcr4a-/- (g) and cxcl12b-/- mutants (h) as compared to wt siblings or age-matched control. Scale bars are 50 µm.
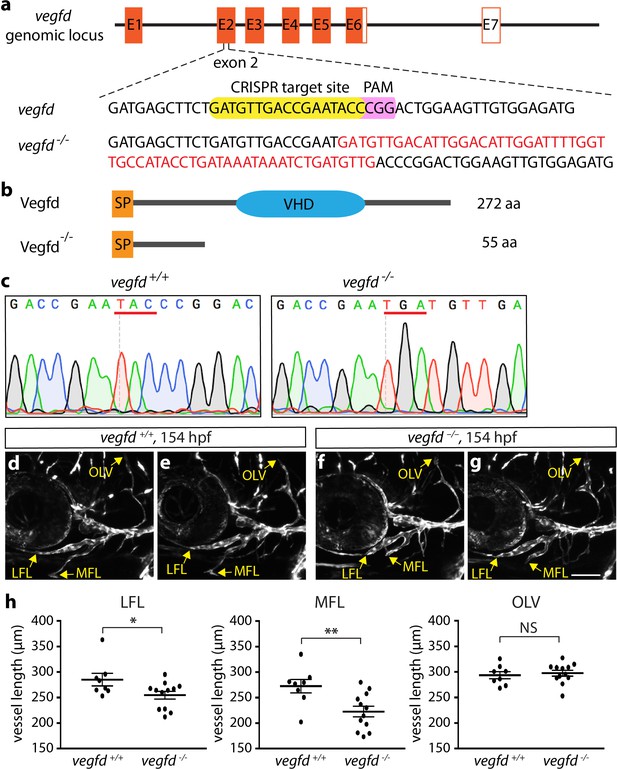
Generation of zebrafish vegfdbns257 mutant allele.
(a) gRNA was designed to target sequences within exon 2 (E2) of the Vegfd protein. The gRNA target site is highlighted in yellow, and the PAM site in purple. Red letters shown in the vegfd allele indicate the CRISPR-induced fifty-nine nucleotide insertion. (b) Schematic diagram of wt and predicted vegfd protein products composed of 272 amino acids (aa) and 55 aa, respectively. SP: signal peptide, VHD: Vegf homology domain. (c) Sequence alignment of part of vegfd exon two from the vegfd+/+ and vegfd -/- allele shows the CRISPR-induced indels leading to the substitution of TAC (tyrosine residue at position 56) for TGA (stop codon) as indicated by red lines. A fifty-nine nucleotide insertion (GATGTTGACATTGGACATTGGATTTTGGTTGCCATACCTGATAAATAAATCTGATGTTG) in vegfd-/- was confirmed by the sequence chromatograms of the PCR products generated using vegfd+/+ and vegfd -/- adult fish genomic DNA as templates (n = 3 for vegfd+/+, n = 8 for vegfd -/-). (d–g) Confocal images showing the facial region of two independent 154 hpf vegfd+/+ (d,e) and vegfd -/- (f,g) larvae carrying the lyve1b reporter. Arrows in each panel point to the vessel terminals of the lateral facial lymphatics (LFL), medial facial lymphatics (MFL), and otolithic lymphatic vessel (OLV). (h) Quantification of average vessel lengths of LFL, MFL, and OLV of 154 hpf vegfd+/+ (n = 8) and vegfd -/- (n = 12) larvae demonstrate significant reduction in average vessel lengths of LFL and MFL. Scale bars are 100 µm.
-
Figure 2—figure supplement 3—source data 1
Quantification of facial lymphatics development in vegfd -/- embryos.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig2-figsupp3-data1-v3.xlsx
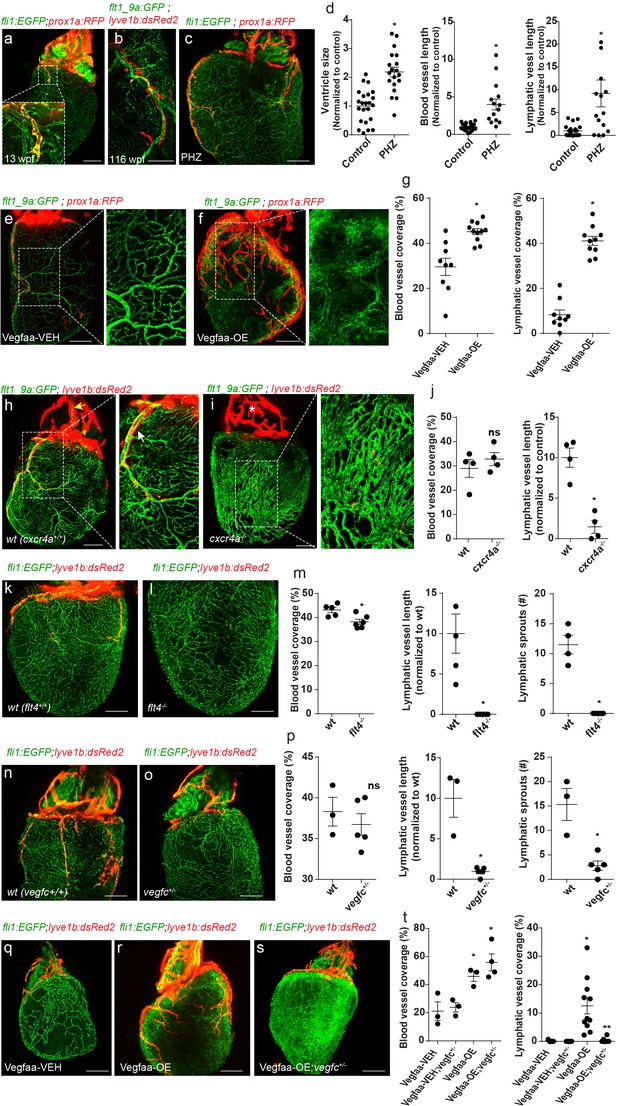
Coronary arteries serve as a scaffold for ventricular lymphatic sprouting.
Insets are magnifications of dashed boxes. (a) 13 wpf (fish size 17 mm) Tg(fli1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) hearts showing ventricular lymphatic sprouting at the base of the OFT (inset) (n = 11). (b) Ventricular lymphatics grow in close proximity to coronary arteries (inset) in 116 wpf (fish size 25–32 mm) Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed2) hearts (n = 15). (c) Heart of 11wpf (fish size 16–22 mm) Tg(fli1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) fish treated with 100 µg/ml Phenylhydrazine hydrochloride (PHZ), showing increased ventricle size and total length of blood and lymphatic vessels following PHZ treatment, quantified in (d) (ncontrol = 21, nPHZ = 15, *p<0.005). (e,f) 12 wpf (fish size 19–22 mm) Tg(βactin2:loxP-mTagBFP-STOP-loxP-vegfaa)pd262; Tg(cmlc2:CreER) fish in the background of Tg(flt1_9a_cFos:GFP);Tg(prox1a:KalTA4-UAS:uncTagRFP). Induction of Vegfaa-OE results in increased blood vessel (insets) and lymphatic vessel coverage, as compared to vehicle (Vegfaa-VEH) treated sibling control, quantified in (g) (nVegfaa-VEH = 9, nVegfaa-OE = 10, *p<0.001). (h–j) Immature coronary plexus, lacking the stereotypical tree-patterning results in nearly absent ventricular lymphatics in cxcr4a-/- hearts of 22 wpf (fish size 25–28 mm) (i), as compared to wt siblings (h), quantified in (j) (nwt = 5, ncxcr4a-/-=6, *p<0.05). (k–m) Ventricular lymphatics are absent in Tg(fli1:EGFP); Tg(lyve1b:dsRed2); flt4-/- hearts (l) at 19–23 wpf (fish size 25–30 mm) as compared to wt siblings (k). (m) Quantification of blood and lymphatic vessel phenotype in flt4 -/- hearts (nwt = 4, nflt4 -/-=5 *p<0.01). (n–p) Tg(fli1:EGFP); Tg(lyve1b:dsRed2); vegfc +/- hearts at 26 wpf (fish size 25–30 mm) display severely defective ventricular lymphatics. (p) Quantification of blood vessel coverage and lymphatic sprout length and number, in vegfc +/- hearts (nwt = 3, nvegfc+/-=5 *p<0.005). (q–t) 12 wpf (fish size 19–22 mm) Tg(βactin2:loxP-mTagBFP-STOP-loxP-vegfaa)pd262; Tg(cmlc2:CreER) fish in the background of Tg(fli1:EGFP);Tg(lyve1b:dsRed2). The increase in lymphatic, but not blood vessel coverage induced by Vegfaa-OE (r), is reversed in vegfc+/- heterozygous animals (s). (t) Quantification of blood and lymphatic vessel coverage in (q–s) (nVegfaa-VEH = 3, nVegfaa-OE = 3, nVegfaa-VEH-blood vessel coverage=3, nVegfaa-OE-lymphatic vessel coverage=10 nVegfaa-OE;vegfc +/- -lymphatic vessel coverage=4, *p<0.01, relative to vehicle treated sibling control, **p<0.001 relative to Vegfaa-OE). Scale bars are 200 µm. Error bars, mean ± S.E.M. All panels show anterior views.
-
Figure 3—source data 1
Quantification of Phenylhydrazine hydrochloride (PHZ)-induced ventricular phenotype.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Quantification of ventricular blood and lymphatic vessel phenotype in VegfAa-OE, flt4 -/-, vegfc+/ - and cxcr4a -/- hearts.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig3-data2-v3.xlsx
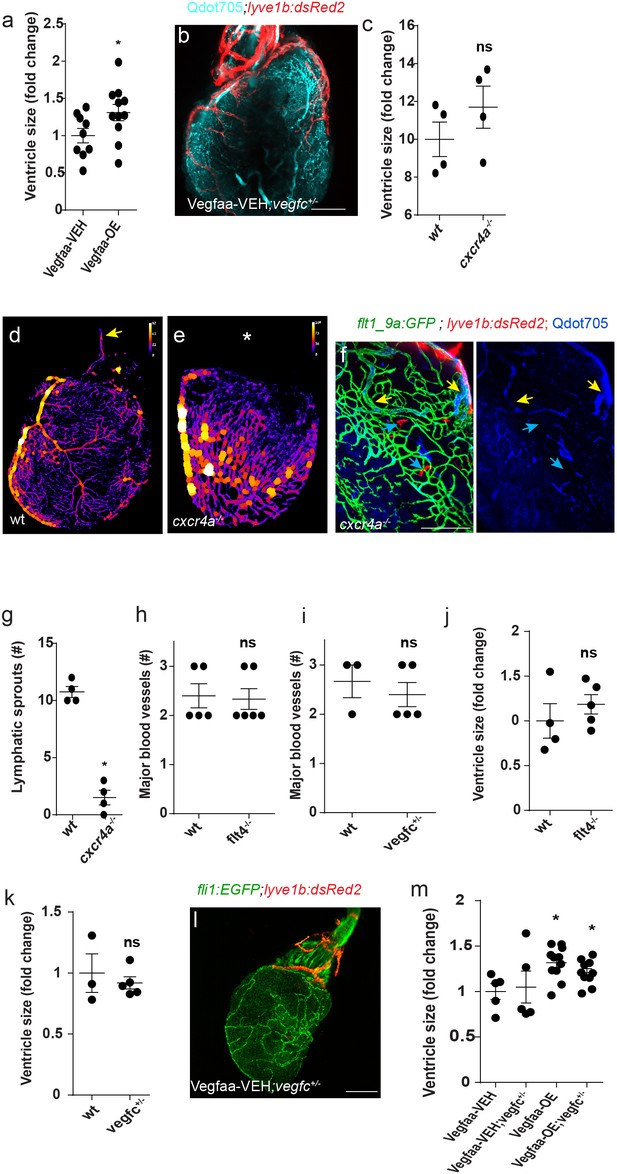
Development of ventricular lymphatics.
Ventricular size is increased following induction of Vegfaa-OE (nVegfaa-VEH = 9, nVegfaa-OE = 10, *p<0.01) (a), but remains unchanged in cxcr4a-/- mutants (c) (nwt = 4, ncxcr4a-/-=4). (b) Lymphatic sprouts (red) follow coronary vessels, labeled by intravascular injection of Qdot705 (cyan), in 12 wpf (fish size 19–22 mm) Vegfaa-OE hearts (n = 4) (d, e) Color-coded maps of vessel thickness show mis-patterned coronary arteries and absent major OFT vessels (e, asterisk) in cxcr4a -/-, as compared to wt siblings (d, yellow arrow). (f) cxcr4a -/- hearts display partially perfused blood coronary vessels (yellow arrows) following intravascular injection of Qdots705 at 25wpf (25–30 mm) (n = 4). (g) The number of ventricular lymphatic sprouts is reduced in 22 wpf (fish size 25–28 mm) Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed2) cxcr4a-/- hearts (nwt = 4, ncxcr4a-/-=4, *p<0.01). (h,i) Quantification of the number of major blood vessels in 19–23 wpf (fish size 25–30 mm) wt sibling and flt4 -/- hearts (h) (nwt = 5, nflt4 = 6) or 24 wpf (fish size 25–30 mm) wt sibling and vegfc+/- hearts (i) (nwt = 3, nvegfc +/-=5). (j,k) Quantification of ventricular size in 19–23 wpf (fish size 25–30 mm) wt sibling and flt4 -/- hearts (j) (nwt = 4, nflt4 = 5) and in 24 wpf (fish size 25–30 mm) wt sibling and vegfc +/- animals (k) (nwt = 3, nvegfc+/-=5). (l) 12 wpf (fish size 19–22 mm) Vegfaa-VEH;vegfc+/- fish in the background of Tg(fli1:EGFP);Tg(lyve1b:dsRed2). (m) Increased ventricular size is not reverted in 12 wpf (fish size 19–22 mm) Vegfaa-OE; vegfc+/- animals (nVegfaa-VEH = 5, nVegfaa-VEH;vegfc+/-=5, nVegfAa-OE-lymphatic vessel coverage=10 nVegfaa-OE;vegfc+/-lymphatic vessel coverage = 9, *p<0.01, relative to Vehicle treated sibling control). Error bars are 200 µm, mean ± S.E.M. All panels show anterior views.
-
Figure 3—figure supplement 1—source data 1
Quantification of ventricular size in Vegfaa-OE, flt4 -/-, vegfc+/ - and cxcr4a -/- hearts.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig3-figsupp1-data1-v3.xlsx
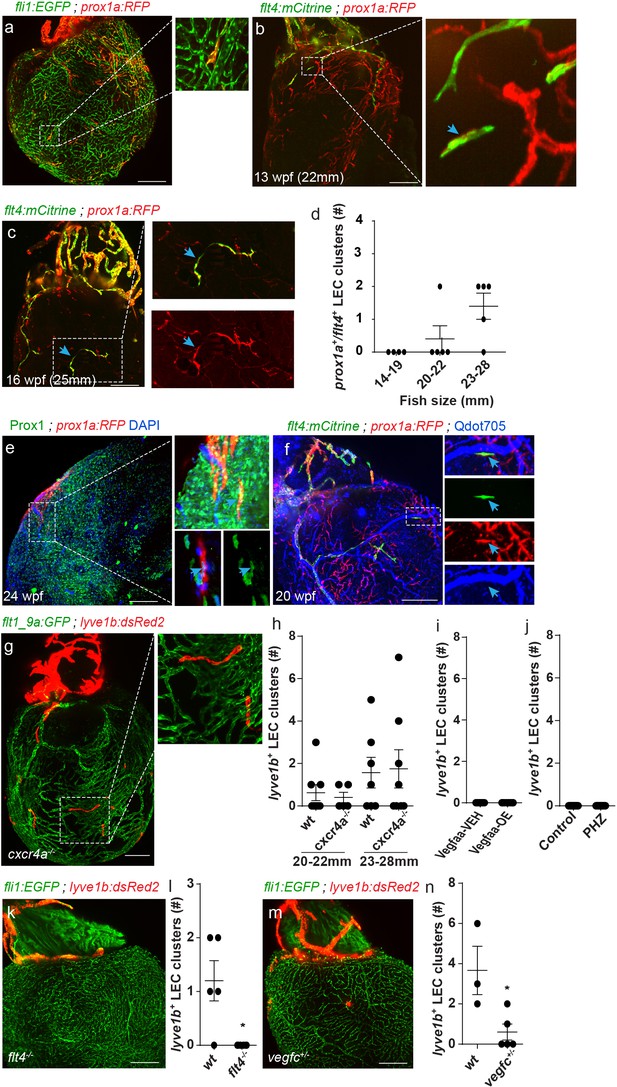
A novel population of isolated LECs is detected in the adult zebrafish heart.
Insets are magnification of dashed boxes. (a) Tg(fli1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) 16 wpf (fish size 23–28 mm) heart showing isolated lymphatic clusters (n = 8). (b,c) Double labeled prox1a+;flt4+ isolated LECs are first detected at ~13 wpf (20–22 mm) (b, arrows) and coalesce to generate isolated capillaries by 16wpf (fish size 25–28 mm) (c, arrows). (d) Quantification of double-labeled prox1a+;flt4+ isolated LECs in the ventricles of 14–28 mm fish (n14-19mm=4, n20-22mm=5, n23-28mm=5). (e) prox1a+ isolated LECs are also labeled by Prox1 antibody (inset, arrow). (f) 20 wpf (fish size 28 mm) double-transgenic prox1a;flt4 hearts demonstrate that isolated LECs are not labeled following intravascular injection of Qdot705 (inset, arrow) (image in f) is an additional view of Figure 2—figure supplement 1c). (g) Isolated LEC clusters develop normally in 22 wpf (fish size 25–30 mm) Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed2); cxcr4a-/- hearts, quantified in (h) (nwt20-22mm=8, ncxcr4a-/- 20-22mm = 6, nwt23-28mm=7, ncxcr4a-/-23-28mm = 8). (i,j) lyve1b+ isolated LEC clusters are not precociously detected in Vegfaa-OE hearts (12.5 wpf, fish size 19–22 mm), in (i) (nveh = 7, nTam = 8) or PHZ treatment (j) (ncontrol = 9, nPHZ = 9) (k). No isolated LECs are detected in 19-23wpf (fish size 23–28 mm) Tg(fli1:EGFP);Tg(lyve1b:dsRed2);flt4 -/- hearts, quantified in (l) (nwt = 5, nflt4-/-=6). (m) Significantly reduced numbers of isolated LECs are detected in Tg(fli1:EGFP);Tg(lyve1b:dsRed2);vegfc+/- animals at 26 wpf (fish size 25–28 mm), quantified in (n) (nwt = 3, nvegfc+/-=5). Scale bars are 200 µm. Error bars, mean ± s.e.m. Anterior view in a-c, e,f,k,m. Posterior view in g.
-
Figure 4—source data 1
Quantification of isolated LECs development in wt, VegfAa-OE, flt4 -/-, vegfc+/ -, cxcr4a -/- and PHZ-treated hearts.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig4-data1-v3.xlsx
Individual Z-stacks through heart shown in Figure 4b confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt4BAC:mCitrine) is in green.
Individual Z-stacks through heart shown in Figure 4g confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt1_9a_cFos:GFP) is in green. Tg(lyve1b:dsRed2) is red.
Individual Z-stacks through heart of a wt sibling of Figure 4g confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt1_9a_cFos:GFP) is in green.Tg(lyve1b:dsRed2) is red.
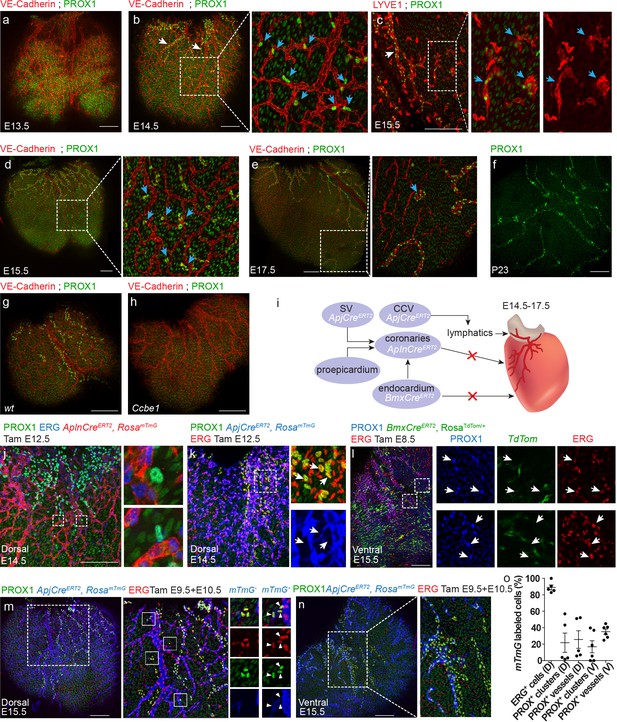
Both lymphangiogenesis and lymph-vasculogenesis contribute to cardiac lymphatic development in mammals.
(a,b) Whole mount confocal images of mouse hearts immunonstained for VE-Cadherin and PROX1. (a) At E13.5 coronary (red) but not lymphatic vessels (green) are present on the ventricle surface (n = 6) (Low levels of PROX1 are detected in cardiomyocytes). (b) In addition to regular lymphatic vessels (white arrows), isolated PROX1+ LECs are observed in close proximity to the coronaries at E14.5 (inset, blue arrows) (n = 6). (c) Whole mount confocal images of E15.5 mouse hearts immunonstained for LYVE1 and PROX1. Isolated PROX1+/LYVE1+ LECs (blue arrows) are not connected to the main lymphatic sprouts (white arrow). (d,e) Whole mount confocal images of mouse hearts immunonstained for VE-Cadherin and PROX1. Isolated LECs gradually expand to form multicellular lumenized structures (inset, arrows) (n = 4). (f) PROX1+ isolated LECs are no longer detected at P23 (n = 4). (g,h) Whole mount confocal images of E15.5 mouse hearts immunonstained for LYVE1 and PROX1. PROX1+ LECs are absent from Ccbe1 mutant hearts (h) (nwt = 3, nCcbe1 = 3). (i) Lineage-tracing strategies for identification of cardiac LEC origins. (j) Dorsal view of AplnCreERT2,RosamTmG heart from embryo dosed with tamoxifen at E12.5 and analyzed at E14.5, showing no AplnCreERT2;PROX1+ LECs. Cre recombination is labeled in red, ECs in blue (Erg) and lymphatics in green (PROX1). Insets are magnification of dashed boxes. (n = 7). (k) Dorsal view of ApjCreERT2,RosamTmG heart, showing that PROX1+ LECs are not labeled by ApjCreERT2 in embryos dosed with tamoxifen at E12.5 and analyzed at E14.5. Cre recombination is marked in blue, ECs in red (ERG) and lymphatics in green (PROX1). Insets are magnification of dashed boxes (n = 4). (l) Ventral view of heart from BmxCreERT2, RosaTdTom embryos dosed with tamoxifen at E8.5 and analyzed at E15.5, showing that PROX1+ LECs are not labeled by BmxCreER. Cre recombination is marked in green, ECs in red (ERG) and lymphatics in blue (PROX1). Inset is magnification of dashed boxes. (n = 5). Dorsal (m) and ventral (n) views of ApjCreERT2,RosamTmG embryos dosed with tamoxifen at E9.5 and 10.5 and analyzed at E15.5, showing PROX1+ isolated LECs and lymphatic vessels, labeled by ApjCreERT2. Cre recombination is marked in blue, ECs in red (ERG) and lymphatics in green (PROX1). (o) Quantification of mTmG labeling shows reduced ApjCreERT2 lineage traced PROX1+ cells as compared to ERG ECs (recombination efficiency). D;Dorsal, V;Ventral (nD = 5, nV = 6) Insets are magnification of dashed boxes. (nDorsal = 6, nVentral = 5). Scale bars are 200 µm.
-
Figure 5—source data 1
Quantification of ApjCre ERT2 lineage traced PROX1+ cells.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig5-data1-v3.xlsx
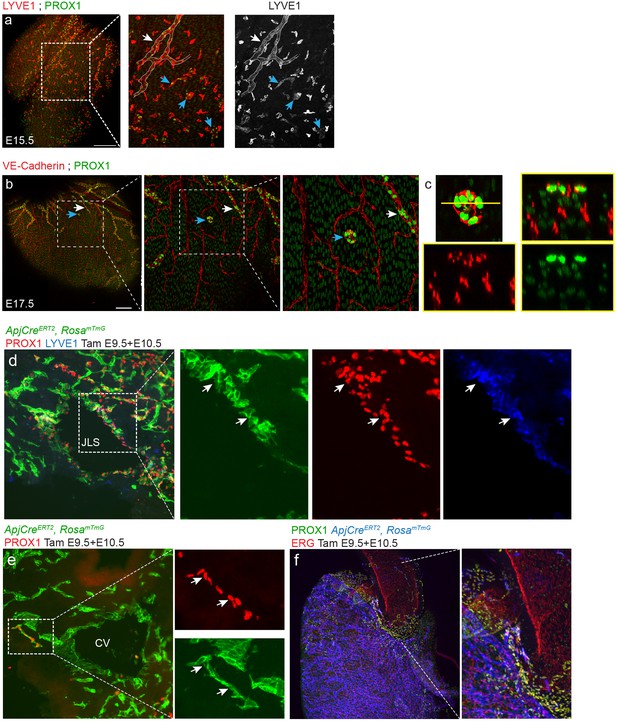
Morphological and lineage analysis of mouse cardiac lymphatics.
(a,b) Whole mount images of embryonic hearts with isolated LEC clusters. (a) Co-staining with the nuclear lymphatic marker PROX1, and the membrane lymphatic marker, LYVE1, revealed that isolated LECs (blue arrow) do not connect to the main lymphatic vessel (white arrow, outline). (b) Isolated LEC clusters (blue arrows) were found as late at E17.5. Insets show increasing magnifications of a cluster that did not contact either VE-Cadherin+ blood ECs or developing lymphatic sprouts (white arrow). (c) An orthogonal view of the cluster highlighted in b) shows that blood vessels from deeper in the tissue do not connect. Scale bars are 200 µm. (d,e) Tissue sections through E11.5 ApjCreERT2,RosamTmG embryos dosed with tamoxifen at E9.5 and 10.5 showing labeling in the jugular lymph sacs (JLS) (green) (d), co-localized with PROX1 (red) and LYVE1 (blue) (inset, arrows). The cardinal vein (CV) is also labeled by mTmG (e) as well as a cluster of PROX1 labeled cell cluster budding from the CV (inset, arrows) (f) Inset is magnification of dashed box. Base of the aorta view of ApjCreER, RosamTmG embryos dosed with tamoxifen at E9.5 and 10.5 and analyzed at E15.5. Cre recombination is marked with GFP (blue), endothelial cells with VE-Cadherin (red) and lymphatics with PROX1 (green). Scale bars are 200 µm.
Individual Z-stacks through LEC cluster shown in Figure 5—figure supplement 1c confirm there are no connections with underlying blood vessel endothelial cells.
VE-Cadherin is in red; PROX1 is in green.
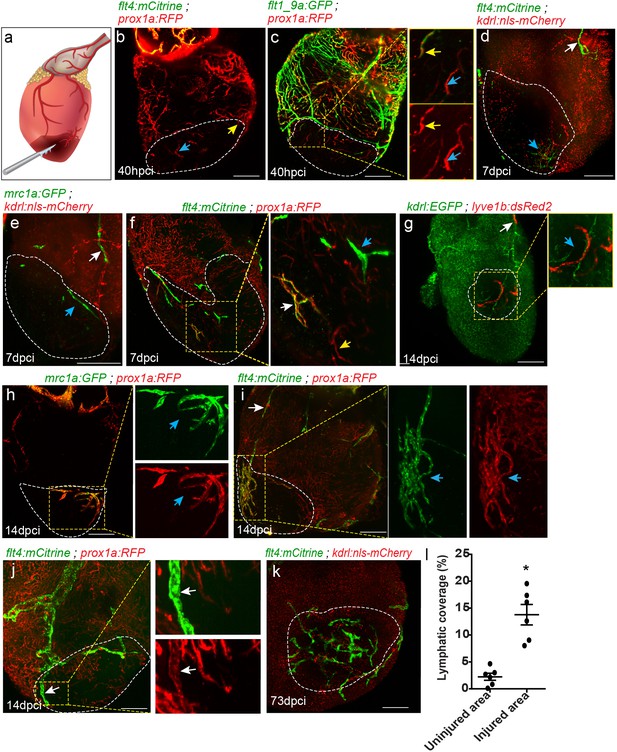
Differential response of cardiac lymphatics to injury.
(a) Diagram depicting the cryoinjury procedure. Injured area is outlined in all images, insets show high-magnification of dashed boxes. (b) flt4;prox1a transgenic hearts at 40 hpci showing prox1a+ sprouts (yellow arrow) and isolated LECs (blue arrow) in the injured area (n = 5). (c) Tg(flt1_9a_cFos:GFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) double labeled coronary sprouts (inset, yellow arrow), as well as prox1a+ isolated LECs (inset, blue arrow) are detected in the injured area at 40 hpci. (d–f) seven dpci injured hearts of (d) Tg(kdrl:nls-mCherry);Tg(flt4BAC:mCitrine) (n = 5), (e) Tg(kdrl:nls-mCherry);Tg(mrc1a:EGFP) (n = 5), and (f) Tg(prox1a:KalTA4-UAS:uncTagRFP);Tg(flt4BAC:mCitrine) (n = 5) fish, with white arrows pointing to OFT-connected ventricular lymphatics, and blue arrows pointing to isolated LECs in the injured area. (g–i) 14 dpci ventricles of (g) Tg(flt1_9a_cFos:GFP;Tg(lyve1b:dsRed2) (n = 8), (h) Tg(prox1a:KalTA4-UAS:uncTagRFP) (n = 5) and (i) Tg(prox1a:KalTA4-UAS:uncTagRFP);Tg(flt4BAC:mCitrine) (n = 5) showing isolated lymphatic sprouts in the injured area (blue arrows), which are not connected to ventricular lymphatics (white arrow) (j) Double labeled prox1a;flt4 ventricular lymphatic sprouts invade the injured area at 14 dpci (inset, white arrows) (n = 3). (k) 73 dpci Tg(kdrl:nls-mCherry);Tg(flt4BAC:mCitrine) heart showing increased lymphatic coverage in the injured vs. uninjured areas of the ventricle, quantified in (l). Error bars, mean ± s.e.m. *p<0.001. Scale bars are 200 µm. Fish size 25–30 mm.
-
Figure 6—source data 1
Quantification of lymphatic coverage 73 dpci.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig6-data1-v3.xlsx
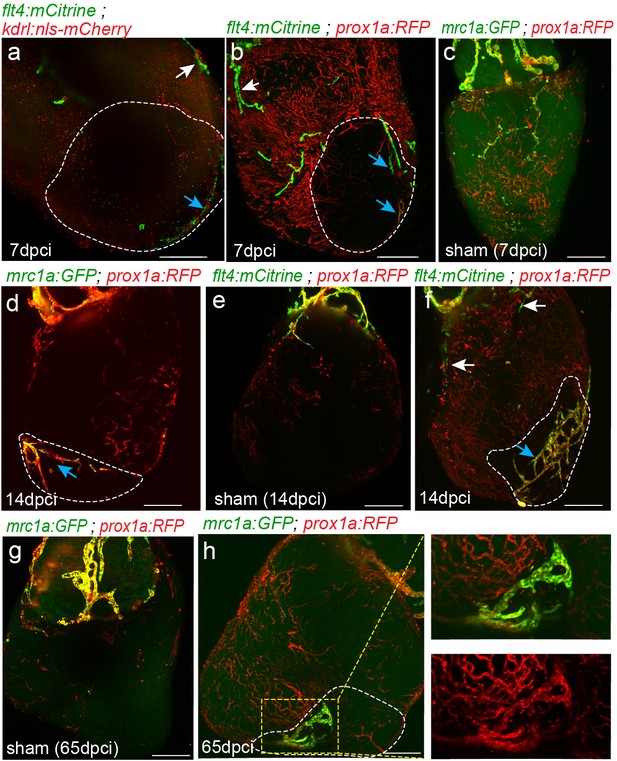
Lymph-vasculogenesis represents the main mechanism of lymphatic formation after cardiac injury.
Injured area is outlined in all images. (a,b) seven dpci ventricles of Tg(kdrl:nls-mCherry);Tg(flt4BAC:mCitrine) (a) and Tg(prox1a:KalTA4-UAS:uncTagRFP); Tg(flt4BAC:mCitrine) (b) showing the opposite side of the hearts presented in Figure 6d,f, confirming the lack of connections between isolated LECs (blue arrows) and OFT originating ventricular lymphatics (white arrows). (c,e) Sham operated Tg(mrc1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) hearts at seven dpci (c) (n = 3), and 65 dpci (g) (n = 2) and Tg(prox1a:KalTA4-UAS:uncTagRFP); Tg(flt4BAC:mCitrine) hearts at 14 dpci (e) (n = 3), showing no changes in lymphatic vasculature. (d,f) Opposite side of the hearts presented in Figure 6h,i, confirming the lack of connections between isolated LECs (blue arrows) and OFT originating ventricular lymphatics (white arrows). (h) Tg(mrc1:EGFP);Tg(prox1a:KalTA4-UAS:uncTagRFP) heart at 65 dpci showing increased lymphatic coverage in the injured vs. the uninjured areas of the ventricle (n = 4). Scale bars are 200 µm. All fish are 25–30 mm.
Individual z-stacks through heart shown in Figure 6f confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt4BAC:mCitrine) is in green. Tg(prox1a:KalTA4-UAS:uncTagRFP) is in red.
Individual z-stacks through heart shown in Figure 6g confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(lyve1b:dsRed2) is in red.
Individual z-stacks through heart shown in Figure 6h confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(mrc1a:EGFP) is green. Tg(prox1a:KalTA4-UAS:uncTagRFP) is in red.
Individual Z-stacks through heart shown in Figure 6—figure supplement 1h confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(mrc1a:EGFP) is green. Tg(prox1a:KalTA4-UAS:uncTagRFP) is in red.
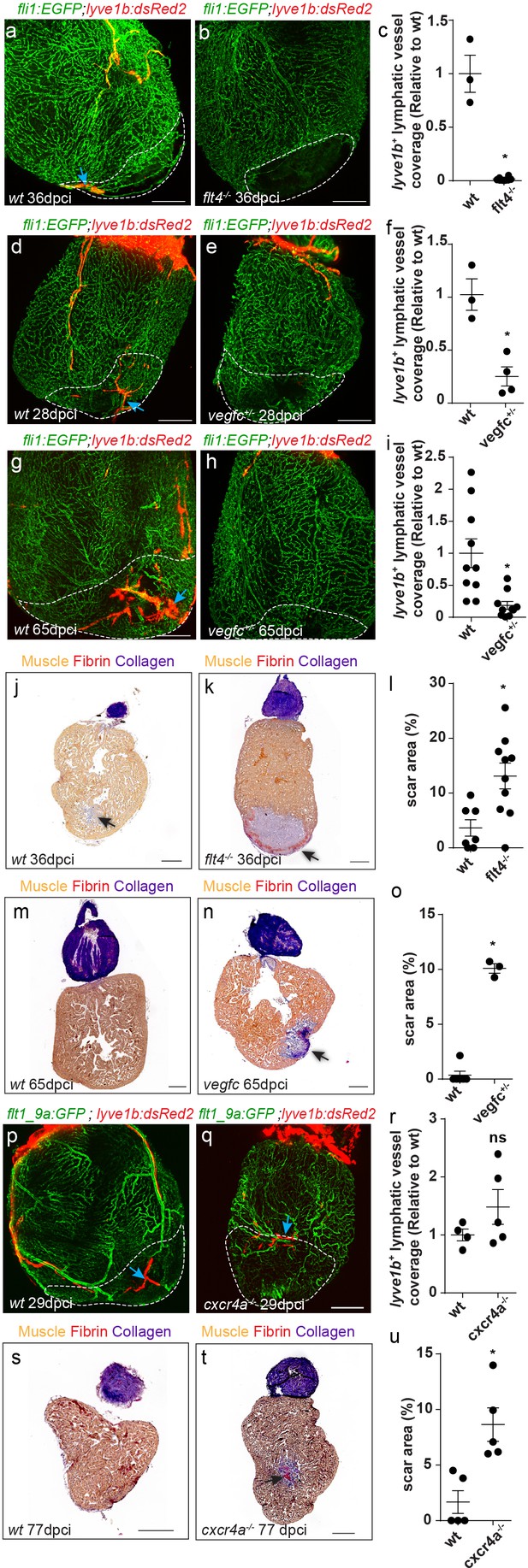
cardiac regeneration is impaired in flt4/vegfc mutant fish.
Injured area is outlined in all images (a,b) lyve1b+ lymphatic capillaries (blue arrows) are detected in the injured area of 36 dpci Tg(fli1:EGFP); Tg(lyve1b:dsRed2) wt sibling (a) but not in flt4-/- hearts (b). (c) Quantification of lyve1b+ lymphatic vessel coverage in the injured area of 35–38 dpi flt4-/- hearts (nwt = 3, nflt4-/-=8, *p<0.001). (d–i) lyve1b+ lymphatic capillaries (blue arrows) are detected in the injured area of 28 dpci (d) and 65 dpci (g) Tg(fli1:EGFP); Tg(lyve1b:dsRed2) wt siblings, but not in vegfc+/- heterozygous fish (e,h). (f,i) Quantification of lyve1b+ lymphatic vessel coverage in the injured area of 21–28 dpi vegfc+/- hearts (nwt = 3 hearts, nvegfc +/-=4, *p<0.005) and 65–66 dpi vegfc +/- hearts (nwt = 10 hearts, nvegfc+/-=10, *p<0.005). (j,k) AFOG-stained sections at 36 dpci showing lack of regeneration in flt4 -/- hearts (k) as compared to wt siblings (j). Collagenous scar is stained in blue, fibrin in red, and cardiac muscle in orange. Black arrow points to scar. (l) Increased scar area (calculated as percent of ventricle) in flt4-/- hearts (nwt = 8 hearts, nflt4-/-=11, *p<0.05). (m,n) AFOG-stained sections at 65 dpci showing lack of regeneration in vegfc+/- hearts (n) as compared to wt siblings (m). Black arrow points to scar. (o) Increased scar area (calculated as percent of ventricle) in vegfc+/- hearts (nwt = 6 hearts, nvegfc+/-=3, *p<0.001). (p,q) lyve1b+ isolated LEC clusters (blue arrows) are normally detected in the injured area of 29 dpci Tg(flt1_9a_cFos:GFP);Tg(lyve1b:dsRed);cxcr4a-/- (q) and wt sibling (p) hearts. (r) Quantification of lyve1b+ lymphatic vessel coverage (relative to wt sibling) in 29 dpi cxcr4a -/- hearts (nwt = 4 hearts, ncxcr4a-/-=5). (s,t) AFOG-stained sections at 77 dpci showing impaired regeneration in cxcr4a-/- mutant (t) as compared to wt siblings (s). Black arrow points to scar. (u) Increased scar area (calculated as percent of ventricle) in cxcr4a-/- mutant hearts (nwt = 5 hearts, ncxcr4a-/-=5, *p<0.005). Scale bars are 200 µm. All fish size are 25–30 mm.
-
Figure 7—source data 1
Quantification of lymphatic coverage and scar area in cryoinjured flt4 -/-, vegfc+/ - and cxcr4a -/- hearts.
- https://cdn.elifesciences.org/articles/44153/elife-44153-fig7-data1-v3.xlsx
Individual Z-stacks through heart shown in Figure 7q confirm there are no connections of isolated lymphatic cluster with the main ventricular lymphatic vasculature.
Tg(flt1_9a_cFos:GFP) is in green. Tg(lyve1b:dsRed2) is in red.

LECs in the injured area do not co-express endocardial, epicardial or myeloid markers.
(a) Section of a Tg(flt4:mCitrine) heart at 7 dpi. Cardiomyocytes are immunostained with anti-MHC antibody (red). Epicardium and injury activated endocardium are immunostained with anti-retinoic acid (RA)-synthesizing enzyme Raldh2 (white). White dotted lines delineate the injured area. Insets show high-magnification of yellow dashed box. Flt4 positive cells in the injured area are not labeled with Raldh2 (b) Tg(flt4:mCitrine);Tg(lyz:dsRed) heart at 6dpi. White dotted lines delineate the injured area. Insets show high-magnification of yellow dashed box. Flt4 positive cells in the injured area do not express the myeloid marker lys. Scale bars are 200µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Danio rerio) | Tg(fli1:EGFP)yl | (Nicenboim et al., 2015) | ZDB-ALT-011017–8 | |
| Strain (Danio rerio) | Tg(lyve1b:dsRed2)nz101 | (Nicenboim et al., 2015) | ZDB-ALT-120723–3 | |
| Strain (Danio rerio) | Tg(gata1a:dsRed)sd2 | (Nicenboim et al., 2015) | ZDB-ALT-051223–6 | |
| Strain (Danio rerio) | TgBAC(prox1a:KalTA4-4xUAS-E1b:uncTagRFP)nim5 | (Nicenboim et al., 2015) | ZDB-ALT-140521–3 | |
| Strain (Danio rerio) | Tg(flt1_9a_cFos:GFP)wz2 | (Nicenboim et al., 2015) | ZDB-ALT-150723–14 | |
| Strain (Danio rerio) | Tg(flt4BAC:mCitrine)hu7135 | (van Impel et al., 2014) | ZDB-ALT-140521–1 | |
| Strain (Danio rerio) | Tg(mrc1a:EGFP)y251 | (Jung et al., 2017) | ZDB-ALT-170717–2 | |
| Strain (Danio rerio) | Tg(kdrl:nls-mCherry)y173 | (Fujita et al., 2011) | ZDB-ALT-110429–4 | |
| Strain (Danio rerio) | Tg(kdrl:EGFP)s843 | (Jin et al., 2005) | ZDB-ALT-050916–14 | |
| Strain (Danio rerio) | cxcr4aum20 | (Siekmann et al., 2009) | ZDB-ALT-091124–1 | |
| Strain (Danio rerio) | cxcl12bmu100 | (Bussmann et al., 2011) | ZDB-ALT-110513–2 | |
| Strain (Danio rerio) | vegfcum18 | (Villefranc et al., 2013) | ZDB-ALT-130718–3 | |
| Strain (Danio rerio) | Tg(βactin2:loxP-mTagBFP-STOP-loxP-vegfaa)pd262; Tg(cmlc2:CreER) | (Karra et al., 2018) | ZDB-ALT-181129–18 | |
| Strain (Danio rerio) | flt4um203 | (Kok et al., 2015) | ZDB-ALT-160721–30 | |
| Strain (Danio rerio) | vegfdbns257 | This paper | N/A | CRISPR/Cas9 generated. Prof. Didier YR Stainier (Max Planck Institute for Heart and Lung Research, Germany) |
| Strain (Mus musculus) | CD1 (wild type) | Charles River Laboratories | Strain# 022 | |
| Strain (Mus musculus) | FVB (wild type) | Charles River Laboratories | Strain# 207 | |
| Strain (Mus musculus) | ApjCreERT2 | (Chen et al., 2014b) | MGI:5689869 | |
| Strain (Mus musculus) | BmxCreERT2 | (Ehling et al., 2013) | MGI:5513853 | |
| Strain (Mus musculus) | AplnCreERT2 | (Liu et al., 2015) | MGI:5637737 | |
| Strain (Mus musculus) | Ccbe1 | (Bos et al., 2011) | N/A | |
| Strain (Mus musculus) | RosamTmG | (Muzumdar et al., 2007) | Stock# 007676 | |
| Strain (Mus musculus) | RosaTdTomato | (Muzumdar et al., 2007) | Stock #007909 | |
| Sequence-based reagent | cxcl12a_F | This paper | PCR primers | CGTAGTAGTCGCT CTGATGG |
| Sequence-based reagent | cxcl12a_R | This paper | PCR primers | TGGGACTGTGTTG ACTGTGGAA |
| Sequence-based reagent | cxcl12b_F | This paper | PCR primers | GGAGCATCCGAGA GATCAAG |
| Sequence-based reagent | cxcl12b_R | This paper | PCR primers | TGTTCTTCAGCTT GGCAATG |
| Sequence-based reagent | Vegfc_F | (Astin et al., 2014) | PCR primers | AAGGGCCCTAACA GAATGTC |
| Sequence-based reagent | Vegfc_R | (Astin et al., 2014) | PCR primers | TTTGAATGAAGGG TGTCAGG |
| Antibody | anti-PROX1 (Rabbit polyclonal) | Abcam | Cat# 11941 | IF(1:700) |
| Antibody | anti-VE-Cadherin (Rat polyclonal) | BD Pharmingen | Cat# 550548 | IF(1:100) |
| Antibody | anti-PROX1 (Goat polyclonal) | R and D Systems | Cat#: AF2727 | IF(1:300) |
| Antibody | anti- ERG (Rabbit monoclonal) | Abcam | Cat#: ab92513 | IF(1:1000) |
| Antibody | anti- LYVE-1 (Rat monoclonal) | eBiosciences | Cat#: 14-0443-80 | IF(1:100) |
| Antibody | Alexa Fluor Conjugated Secondary Antibodies (488,594,633,635,647) | Life Technologies | N/A | IF(1:250) |
| Chemical compound, drug | Atenolol | Sigma Aldrich | A7655 | |
| Chemical compound, drug | Phenylhydrazine hydrochloride (PHZ) | Sigma Aldrich | 78690 | |
| Chemical compound, drug | 4-hydroxytamoxifen | Sigma Aldrich | H7904 | |
| Commercial assay or kit | Qtracker705 | Invitrogen | Q21061MP | |
| Commercial assay or kit | Acid Fuchsin Orange-G (AFOG) | DIAPATH | 010307 | |
| Software, algorithm | Angiotool | (Zudaire et al., 2011) | N/A | |
| Software, algorithm | Image J | NIH (https://www.nih.gov/ij/) | N/A |





