tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay
Figures
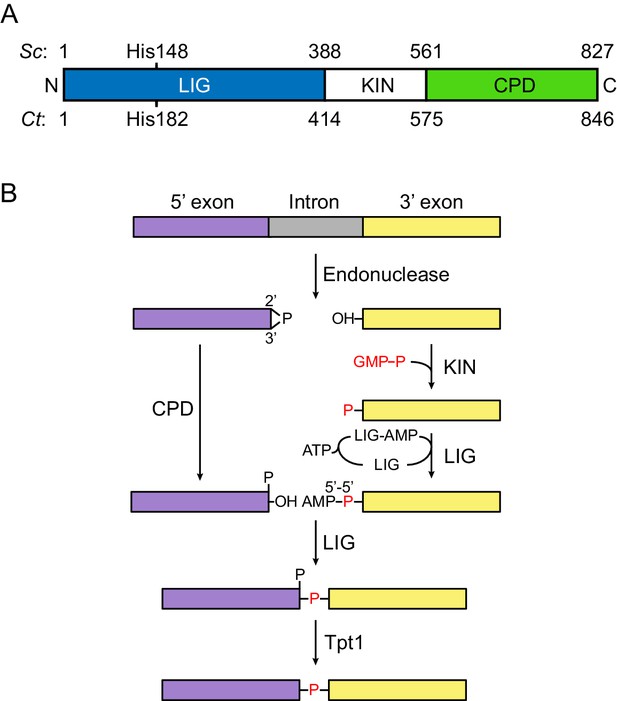
Fungal tRNA ligase Trl1.
(A) Domain organization of Trl1. The ligase/adenylyltransferase domain (LIG) is shown in blue, the polynucleotide kinase domain (KIN) in white, the cyclic phosphodiesterase domain (CPD) in green. The residue numbering refers to the domain boundaries of Trl1 from Saccharomyces cerevisiae (Sc) and Chaetomium thermophilum (Ct). The relative position of the mutated histidine in the UPR mutant is indicated. See Figure 1—figure supplement 1 for a sequence alignment of Trl1-LIG from both species. (B) Trl1-mediated non-conventional RNA splicing mechanism. The RNA is cleaved at the exon-intron junctions by an endonuclease (SEN or Ire1), which leaves a 2’,3’-cyclic phosphate end on the 5’ exon (purple) and a 5’-OH on the 3’ exon (yellow). After removal of the intron (gray), the exon ends are first modified (‘healed’) by the CPD and KIN domains and finally ligated (‘sealed’) by the LIG domain. Exon ligation progresses via a covalent ligase-AMP intermediate (LIG-AMP) and a 5’−5’ RNA-adenylate. The residual 2’-phosphate at the splice site is then removed by a separate enzyme, the 2’-phosphotransferase Tpt1.
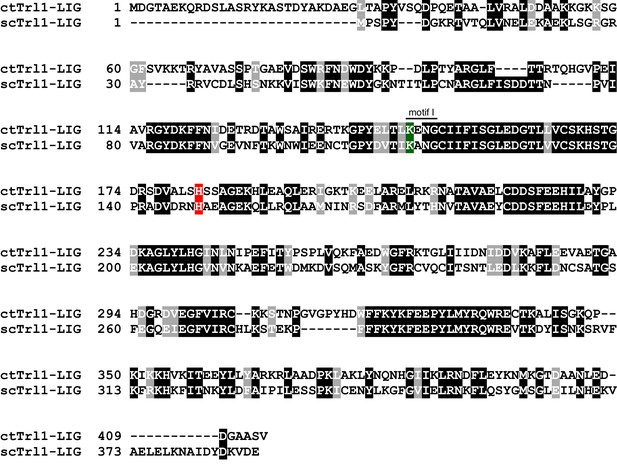
Sequence alignment between scTrl1 and ctTrl1.
Sequence alignment of Trl1-LIG from Chaetomium thermophilum (ct) and Saccharomyces cerevisiae (sc). White letters with gray or black background indicate similarity or identity, respectively. The active site lysine, which is part of the highly conserved motif I, and the histidine that is mutated to tyrosine in the UPR mutant are highlighted in green and red, respectively.
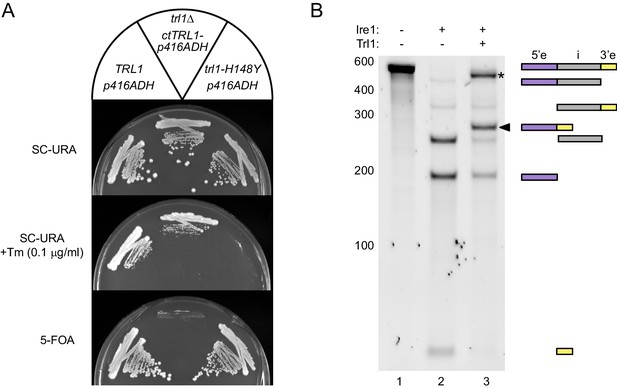
Functional complementation by thermophilic ctTrl1.
(A) Functional complementation in S. cerevisiae trl1Δ by ctTrl1. Expression of ctTrl1 from a plasmid (URA3 selection) permitted growth of a trl1Δ strain under normal growth conditions (SC-URA) and ER stress (SC-URA +Tm). Counterselection against the URA3-containing plasmid using 5’-fluoroorotic acid (5-FOA) confirmed the essential function of Trl1. The respective TRL1 genotype and plasmid are indicated in the key on top. (B) In vitro splicing of a HAC1-derrived RNA substrate was analyzed by denaturing urea-PAGE. The input RNA (lane 1) was first cleaved by scIre1 yielding three dominant bands for the separated exons and the intron (lane 2). Faint bands corresponding to single cleaved RNA were observed. Addition of ctTrl1 completed the splicing reaction resulting in covalently ligated exons (lane 3, marked by a closed arrowhead). An additional ligation product is marked with an asterisk. RNase H digestion identified the band as circularized tandem HAC1 intron RNA (see Figure 2—figure supplement 1). The position on the gel and relative length of the RNA species is depicted by icons on the right using the same color code as in Figure 1B.
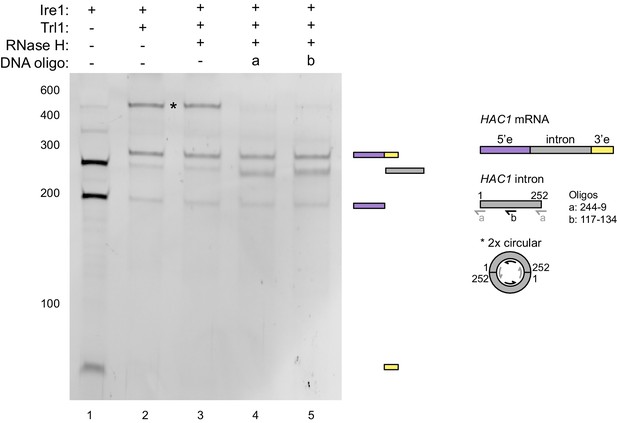
RNase H assay.
In vitro transcribed HAC1U-508 RNA was cleaved by Ire1 (lane 1) and subsequently ligated by Trl1 (lane 2). The ligation products were hybridized with DNA oligonucleotides a (lane 4) or b (lane 5) and treated with RNase H. In lane 3, no oligonucleotide was added. The RNA products after each step were analyzed on a denaturing polyacrylamide gel and visualized by SYBR Gold staining. The band in question (marked with an asterisk) disappeared upon RNase H digestion of the RNA/DNA hybrids and a band at the position of the severed intron (252 nt) reappeared. Based on this result and the position on the gel, the marked band represents most likely two introns that were circularized by two intermolecular ligation events at their 5’ and 3’ ends. Hybridization positions of the DNA oligonucleotides on a linear HAC1 intron and the putative circular concatemer of two introns are schematically described in the right.
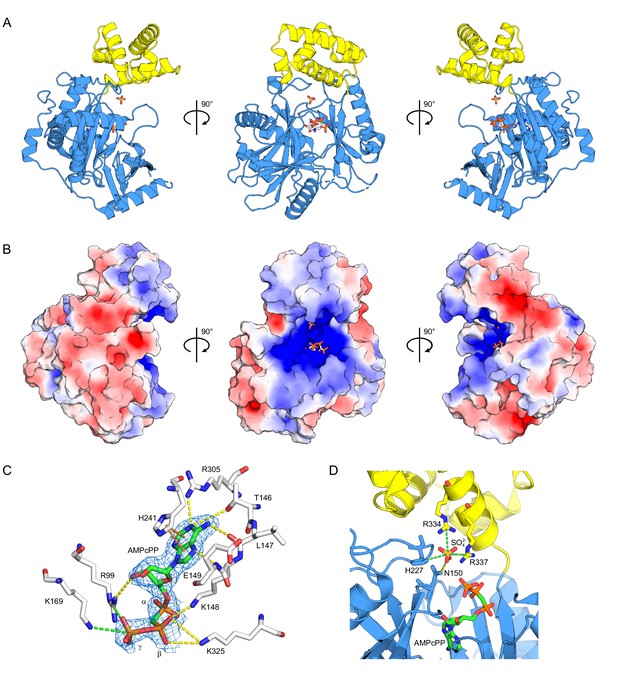
Structure of ctTrl1-LIG.
(A) Cartoon representation of the overall structure of ctTrl1-LIG from three views. The adenylyltransferase domain is colored in blue and the C-terminal domain in yellow. AMPcPP and the sulfate ion near the active site are depicted as sticks. (B) Electrostatic surface potential map of ctTrl1-LIG. Positive potential is represented in blue and negative potential in red. (C) ATP binding pocket with bound AMPcPP. The interacting amino acid residues and AMPcPP are shown as sticks. Atomic contacts between ctTrl1-LIG and AMPcPP are indicated by dashed lines, with yellow for hydrogen bonds, orange for π-stacking and green for salt bridges. The electron density around AMPcPP (blue mesh) is extracted from a composite omit map (2mFo-DFc), contoured at 1 σ. (D) Sulfate ion binding near the active site. Amino acids, AMPcPP and the sulfate ion are shown as sticks. C atoms are colored according to the respective domains (see A) in blue (adenylyltransferase domain) or yellow (C terminal domain). Atomic contacts are depicted as in C.
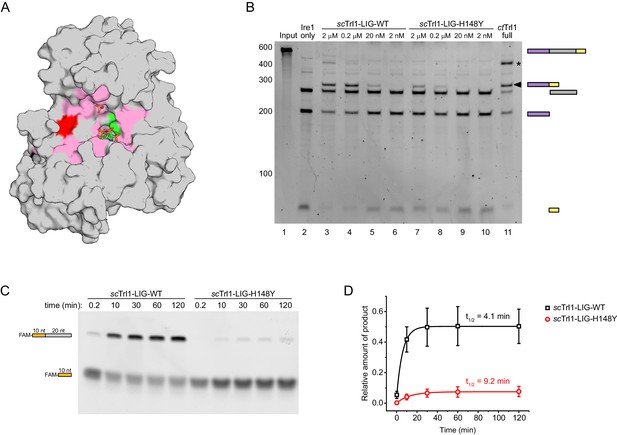
The UPR mutant trl1-H148Y compromises RNA ligation kinetics of Trl1.
(A) Functionally important residues of Trl1-LIG. The structure of ctTrl1-LIG is depicted as surface model, AMPcPP and the sulfate ion are shown as sticks. The active site Lys is depicted in green, His182 (equivalent to His148 in S. cerevisiae) in red and residues that have been previously identified as essential by mutagenesis in pink (Wang and Shuman, 2005). (B) In vitro splicing of a HAC1-derrived RNA substrate was analyzed by denaturing urea-PAGE. The input RNA (lane 1) was first cleaved by scIre1 yielding three dominant bands for the separated exons and the intron (lane 2). Ligation efficiency was compared by titrating purified scTrl1-LIG-WT (lanes 3–6) or scTrl1-LIG-H148Y (lanes 7–10) to the splicing reaction. Full-length ctTrl1 was used as positive control (lane 11). An additional ligation product is marked with an asterisk. RNase H digestion identified the band as circularized HAC1 intron RNA (see Figure 2—figure supplement 1). (C) Time-course denaturing urea-PAGE of RNA oligonucleotide ligation by WT or H148Y mutant scTrl1-LIG. The 5’ fluorescein (FAM)-labeled 10 nt HAC1 5’ exon oligonucleotide (depicted in orange, lower band) and its ligation product (upper band) with a 20 nt HAC1 3’ exon oligonucleotide (depicted in gray) was monitored using fluorescence detection. A representative gel out of three replicates is shown. (D) Quantification of the RNA ligation assay in C. The relative amounts of ligated product in the presence of scTrl1-LIG-WT (black squares) or scTrl1-LIG-H148Y (red circles) were fitted to a first-order model to determine the depicted half-lives. Values represent mean and standard deviation of technical replicates (n = 3).
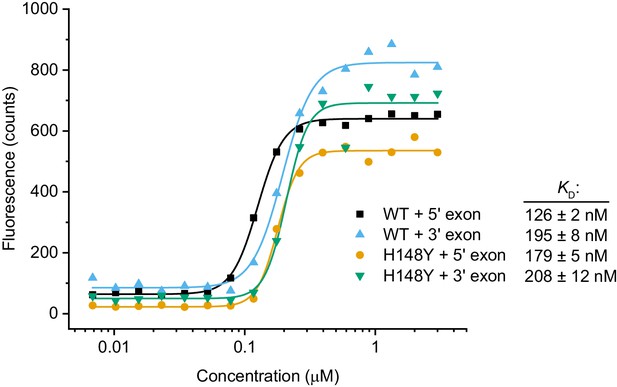
RNA binding assay.
Cy5-labeled RNA oligonucleotides derived from the HAC1 sequences of the 5’ and 3’ exon splice sites were incubated with a dilution series of scTrl1-LIG-WT or -H148Y. The concentration-dependent changes in Cy5 fluorescence were fitted to a Hill model of binding. The calculated apparent dissociation constants (KD) are depicted next to the respective ligase/RNA combination.
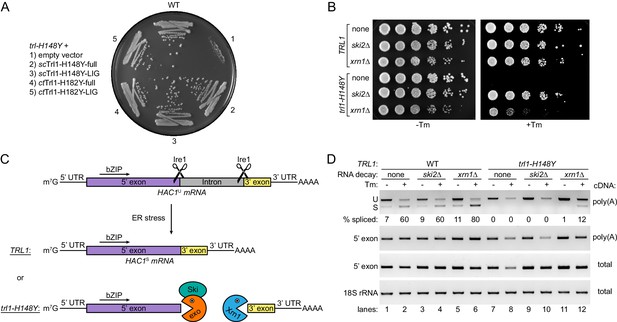
HAC1 mRNA splicing competes with general RNA decay pathways.
(A) Overexpression of various Trl1 constructs rescued the growth phenotype of trl1-H148Y. The constructs from S. cerevisiae harbored the H148Y mutation of the trl1-H148Y allele, whereas those from C. thermophilum harbored the equivalent H182Y mutation. Both the full-length proteins and the isolated ligase domains restored growth of the UPR mutant strain when streaked on tunicamycin-containing YPD plates. (B) Serial five-fold dilutions of wild-type and trl1-H148Y cells with wild-type or abrogated (ski2Δ or xrn1Δ) RNA decay pathways were spotted on YPD plates without (-Tm) or with (+Tm) 0.1 μg/ml tunicamycin and incubated at 30°C for 2 days. (C) Model of HAC1 mRNA splicing and the impact of RNA decay pathways on degradation of cleaved HAC1 exons. Ire1 cleaves unspliced HAC1 mRNA (HAC1U) at the non-conventional splice sites upon ER stress. In wild-type yeast cells (TRL1), exon-exon ligation by Trl1 is the predominant reaction yielding spliced HAC1 mRNA (HAC1S). In the context of a kinetically compromised tRNA ligase (trl1-H148Y), the cleaved HAC1 exons are degraded by exonucleolytic RNA decay pathways. The capped (m7G) 5’ exon is susceptible to degradation in 3’-to-5’ direction by the RNA exosome (exo)/Ski complex (Ski); the 3’ exon is degraded from its 5’ end by Xrn1. (D) RT-PCR analysis of HAC1 mRNA splicing in the same strains as in B without and with tunicamycin (Tm). The top panel shows priming outside the exon-intron junctions to distinguish unspliced (U) from spliced (S) HAC1 mRNA. The relative amount of HAC1S (in %) is indicated below each lane. Both middle panels show the results for priming of the 5’ exon. 18S rRNA was used as a control (bottom panel). The method of cDNA production is indicated on the right of each panel.
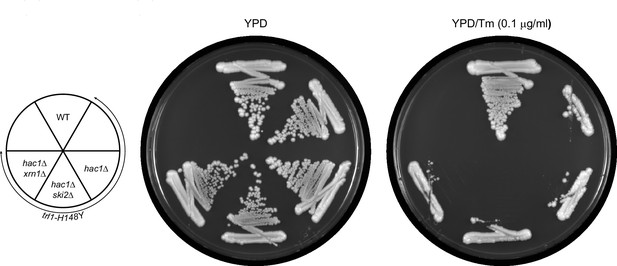
The rescue of UPR-deficiency in trl1-H148Y is dependent on HAC1.
The indicated strains (key in the left) were tested for growth on YPD plates without (left plate) and with (right plate) tunicamycin (Tm).
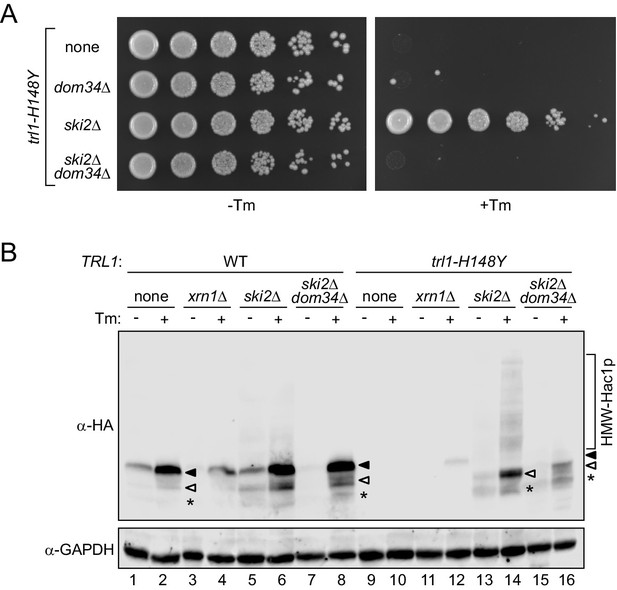
Ribosome rescue allows UPR signaling from the HAC1 5’ exon fragment.
(A) Evaluating the role of ribosome recycling on the ski2Δ-dependent growth rescue of trl1-H148Y cells under ER stress. Serial five-fold dilutions of trl1-H148Y cells with the indicated deletions of SKI2 and/or DOM34 were spotted on YPD plates without (-Tm) or with (+Tm) 0.1 μg/ml tunicamycin and incubated at 30°C for 2 days. (B) Lysates of the indicated strains were prepared and immunoblotted for 3xHA-Hac1p and GAPDH as loading control. Full-length Hac1p is indicated by a closed arrowhead, truncated Hac1p by an open arrowhead. An even shorter minor third band is marked with an asterisk. The high molecular weight smear/banding pattern is indicated as HMW-Hac1p.
Tables
Data collection and refinement statistics.
https://doi.org/10.7554/eLife.44199.007| Datasets: | High-resolution (remote) | Se SAD phasing (peak) |
|---|---|---|
| Data collection | ||
| Beamline | APS 23ID-D | APS 23ID-D |
| Wavelength (Å) | 1.0332 | 0.9793 |
| Space group | P212121 | P212121 |
| Cell dimensions a, b, c (Å) α, β, γ (°) | 49.73, 56.58, 173.88 90, 90, 90 | 49.63, 56.46, 173.87 90, 90, 90 |
| Resolution (Å) | 37.74–1.90 (1.94–1.90) | 49.63–2.50 (2.60–2.50) |
| Total reflections | 159936 (10248) | 175374 (20071) |
| Unique reflections | 39302 (2495) | 17561 (1930) |
| Multiplicity | 4.1 (4.1) | 10.0 (10.4) |
| Completeness (%) | 99.3 (99.1) | 99.1 (98.1) |
| I/σ(I) | 10.3 (1.2) | 21.9 (7.3) |
| CC(1/2) | 0.998 (0.793) | 0.999 (0.982) |
| Rpim | 0.036 (0.575) | 0.024 (0.090) |
| Phasing | ||
| Number of Se sites | 2 | |
| Figure of merit | 0.64 | |
| Refinement | ||
| Rwork/Rfree | 0.170/0.214 | |
| Number of atoms | 3457 | |
| Wilson B factor (Å) | 33.8 | |
| RMS deviations Bonds (Å) Angles (°) | 0.007 1.386 | |
| Ramachandran plot % favored % allowed % outliers | 97.91 1.83 0 | |
| PDB ID | 6N67 |
-
Values in parentheses refer to the highest resolution shell.
The Rfree set consists of 5% randomly chosen data excluded from refinement.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Chaetomium thermophilum) | TRL1 | Chaetomium thermophilum genome resource | Ct:CTHT_0034810 | |
| Gene (Saccharomyces cerevisiae) | TRL1 | Saccharomyces Genome Database | SGD:S000003623 | |
| Gene (Saccharomyces cerevisiae) | IRE1 | Saccharomyces Genome Database | SGD:S000001121 | |
| Gene (Saccharomyces cerevisiae) | HAC1 | Saccharomyces Genome Database | SGD:S000001863 | |
| Antibody | anti-HA (mouse monoclonal) | Sigma | Sigma:H3663; RRID:AB_262051 | (1:2000) |
| Antibody | anti-GAPDH (rabbit polyclonal) | abcam | abcam:ab9485; RRID:AB_307275 | (1:1000) |
| Antibody | goat anti-mouse (goat polyclonal, HRP conjugate) | Promega | Promega:W4021; RRID:AB_430834 | (1:10000) |
| Antibody | goat anti-rabbit (rabbit polyclonal, HRP conjugate) | Promega | Promega:W4011; RRID:AB_430833 | (1:10000) |
| Recombinant DNA reagent | HAC1U-508-pBSK- | PMID:9323131 | lab archive:pPW0386; paper:pCF187 | HAC1 in vitro transcript (508 nucleotides) |
| Sequence-based reagent | Cy5-5' exon HAC1 RNA oligonucleotide | IDT | 5’-Cy5-CGUAAUCCAG-3’-PO4 | |
| Sequence-based reagent | Cy5-3' exon HAC1 RNA oligonucleotide | IDT | 5’-PO4-AAGCGCAGUC-Cy5 | |
| Sequence-based reagent | FAM-5' exon HAC1 RNA oligonucleotide | IDT | 5’-FAM-CGUAAUCCAG-3’-PO4 | |
| Sequence-based reagent | 3' exon HAC1 RNA oligonucleotide | IDT | 5’-PO4-AAGCGCAGUCAGGUUUGAAU-3’ | |
| Sequence-based reagent | oligo a for RNase H assay | IDT | TAATCACGGCGGACAGTA | |
| Sequence-based reagent | oligo b for RNase H assay | IDT | TTGAAGGTACTTTAACCG | |
| Sequence-based reagent | oligo-(dT)12-18 | Thermo Fisher Scientific | Thermo Fisher Scientific:18418012 | |
| Sequence-based reagent | random hexamers | Thermo Fisher Scientific | Thermo Fisher Scientific:N8080127 | |
| Peptide, recombinant protein | ctTrl1 [full length] | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | ctTrl1-LIG | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | scTrl1-LIG-WT | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | scTrl1-LIG-H148Y | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | scTrl1-CPD | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | scIre1-KR32 | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | RtcA | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | His10-HRV-3C protease | This paper | purified from E. coli BL21-RIL cells | |
| Peptide, recombinant protein | T4 polynucleotide kinase (3’ phosphatase minus) | New England BioLabs | New England BioLabs:M0236S | |
| Peptide, recombinant protein | Proteinase K | Thermo Fisher Scientific | Thermo Fisher Scientific:25530049 | |
| Peptide, recombinant protein | RNAse H | New England BioLabs | New England BioLabs:M0297S | |
| Peptide, recombinant protein | SuperScript II reverse transcriptase | Thermo Fisher Scientific | Thermo Fisher Scientific:18064014 | |
| Chemical compound, drug | AMPcPP | Sigma | Sigma:M6517 | |
| Chemical compound, drug | 4μ8C | Matrix Scientific | Matrix Scientific:037985; CAS:14003-96-4 | |
| Chemical compound, drug | Tunicamycin | Sigma | Sigma:T7765 | |
| Chemical compound, drug | SYBR Gold nucleic acid stain | Thermo Fisher Scientific | Thermo Fisher Scientific:S11494 |
Additional files
-
Supplementary file 1
List of plasmids.
- https://doi.org/10.7554/eLife.44199.013
-
Supplementary file 2
Yeast strains (generated in this study).
- https://doi.org/10.7554/eLife.44199.014
-
Supplementary file 3
RT-PCR primers and experimental settings.
- https://doi.org/10.7554/eLife.44199.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44199.016





