Primed Track, high-fidelity lineage tracing in mouse pre-implantation embryos using primed conversion of photoconvertible proteins
Figures
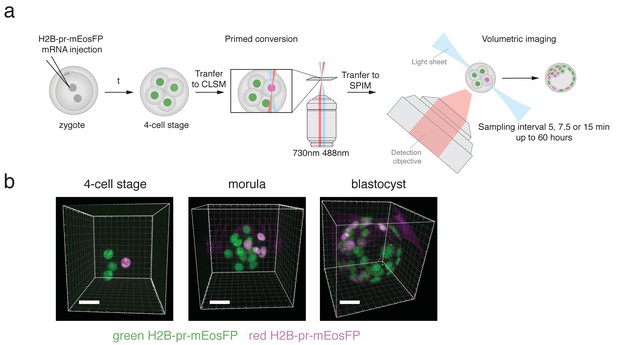
H2B-pr-mEosFP injected embryos develop to the blastocyst stage.
(a) Experimental setup: Zygotes are injected with H2B-pr-mEosFP mRNA. At the 4-cell stage confined primed conversion of a single nucleus is performed using intersecting 488 nm and 730 nm lasers. The embryos are transferred to an inverted SPIM for non-invasive imaging of their development up to the blastocyst stage. Images are taken every 5, 7.5 or 15 min. (b) Embryos injected with mRNA encoding H2B-pr-mEosFP and converted at the four-cell-stage develop normally and maintain visibility of the red label up to the early blastocyst stage. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (magenta). N ≥ 200 embryos out of ≥10 independent experiments. Scale bar, 20 µm.

Embryos expressing H2B-pr-mEosFP develop normally.
https://doi.org/10.7554/eLife.44491.007
Visualizing the lineage of a single cell up to the blastocyst stage using primed converted at the 4-cell stage.
https://doi.org/10.7554/eLife.44491.008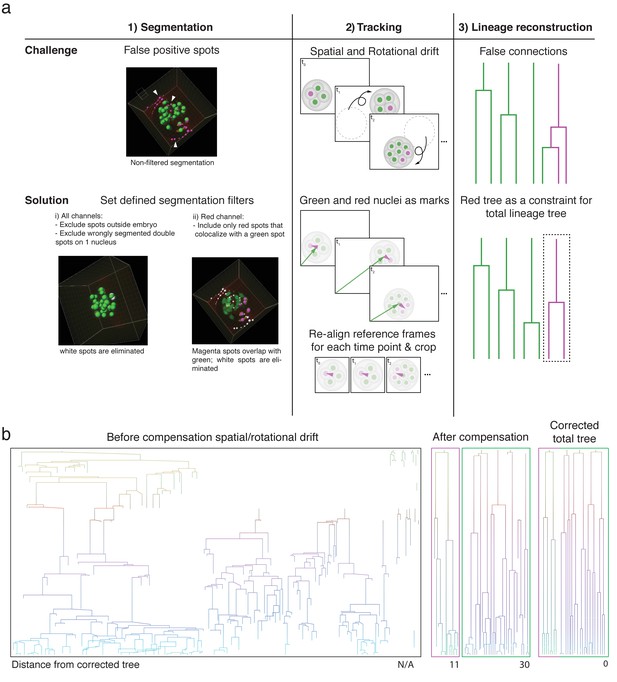
Primed track results in efficient lineage reconstruction of embryos with high spatial and rotational drift.
(a) Overview of the pipeline used for reliable automated segmentation, tracking, and lineage tracing of the imaged embryos; (1) Segmentation: low thresholds are used for the spot detection in both the green and red channel to enable detection of dimmer cells at later developmental time points. Incorrectly segmented spots are excluded by defined filters: (i) exclusion of spots outside of a defined radius of the embryo, (ii) replacement of incorrectly segmented double spots by one spot per one nucleus, and (ii) exclusion of red spots that do not colocalize with green nuclear spots. (2) Tracking: Spatial drift as well as rapid embryo rotation complicates tracking nuclei over prolonged time windows. The segmented nuclei are used for defining reference frames based on the center of mass of the green nuclei and the orientation of the red nuclei. The alignment of the references frames of each time point compensates the spatial and rotational drifts. (3) Lineage tracing: Automated lineage tree reconstruction can make false connections when cells are dividing. By separating the calculation of the lineage trees in the photoconverted red channel from the green channel, the less complex datasets for each channel result in more consistent lineage tracing. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (magenta) overlaid with segmentation results (green and Magenta spheres); Scale bar, 20 µm (b) Lineage trees from the same embryo (corresponding to Videos 1 and 4) reconstructed from segmented nuclei before correction for rotational and translational drift (left), after correction for rotational and translational drift for the red channel (second left), after correction for rotational and translational drift for the green channel minus the spots that colocalize with the red spots (second right), and after final manual lineage reconstruction (right). The embryo was imaged every 15 min.
-
Figure 2—source data 1
Summary of challenges with state-of-the-art segmentation and tracking tools.
- https://doi.org/10.7554/eLife.44491.013
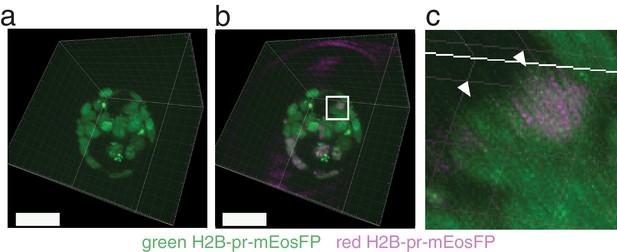
Dual labeling facilitates segmentation in dense environments.
https://doi.org/10.7554/eLife.44491.010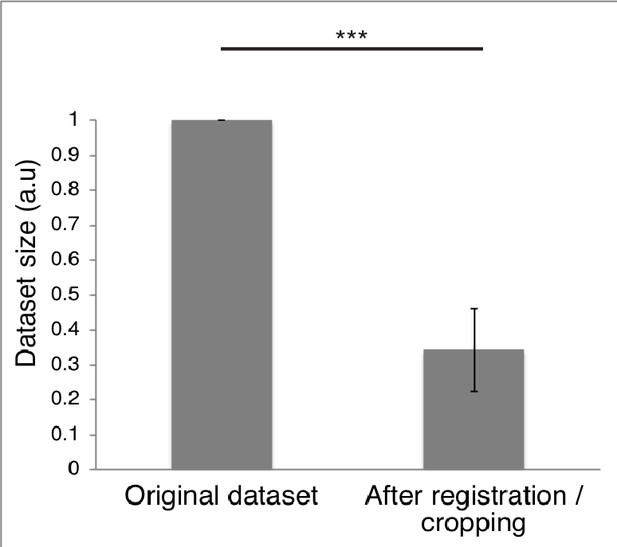
Embryo dataset size before and after registration.
https://doi.org/10.7554/eLife.44491.011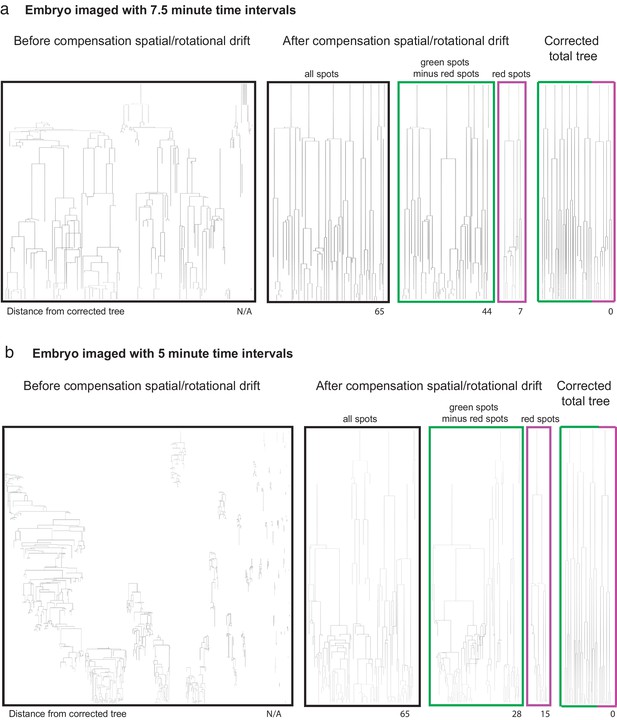
Comparison of lineage tracing results.
https://doi.org/10.7554/eLife.44491.012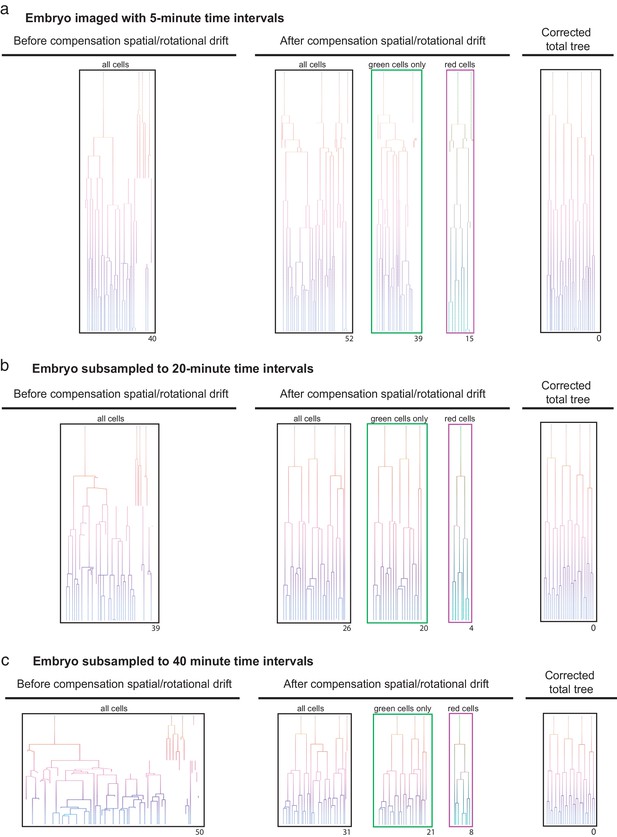
Efficient lineage reconstruction using primed Track is still achieved at large imaging time intervals An originally non-rotating embryo imaged with 5 min.
(a) time intervals were subsampled to 20 min (b) and 40 min (c) time intervals. Lineage tracing was performed on the non-processed dataset as well as after correction for spatial and rotational drift using the presented approach. The numbers displayed below each lineage tree indicate the distance to the final correct lineage tree.
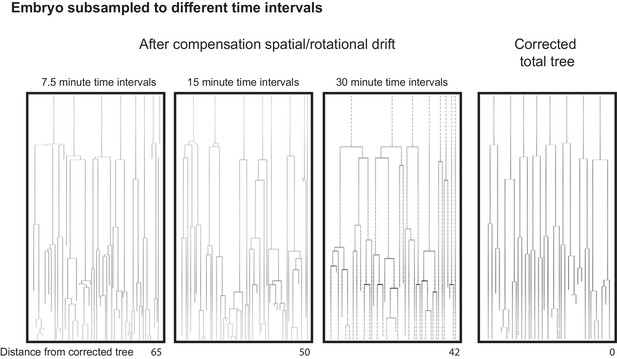
Efficient lineage reconstruction using primed Track is still achieved at large imaging time intervals in a rotating embryo.
https://doi.org/10.7554/eLife.44491.015Videos
Video of a developing embryo before drift correction imaged every 15 mintes.
Timelapse video of an example embryo, which shows strong spatial and rotational drift before drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Scale bars, 15 µm; framerate: one frame every 15 min.
Video of another developing embryo before drift correction images every 7.5 min.
Timelapse video of an example embryo, which shows strong spatial and rotational drift before drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Scale bars, 10 µm; framerate: one frame every 7.5 min.
Video of another developing embryo before drift correction images every 5 min.
Timelapse video of an example embryo, which shows strong spatial and rotational drift before drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Scale bars, 30 µm; framerate: one frame every 5 min.
Video of a developing embryo (same as in Video 1) after drift correction.
Timelapse video of the example embryo from Video 1 after drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Corresponding lineage trees are displayed in Figure 2d. Scale bars, 15 µm; framerate: one frame every 15 min.
Video of a developing embryo (same as in Video 2) after drift correction.
Timelapse video of the example embryo from Video 2 after drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Corresponding lineage trees are displayed in Figure 2—source data 1. Scale bars, 10 µm; framerate: one frame every 7.5 min.
Video of a developing embryo (same as in Video 3) after drift correction.
Timelapse video of the example embryo from Video 2 after drift correction. pr-mEosFP fluorescence (green) and primed converted pr-mEosFP fluorescence (red). Corresponding lineage trees are displayed in Figure 2—source data 1. Scale bars, 30 µm; framerate: one frame every 5 min.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44491.019





