Nedd4 E3 ligase and beta-arrestins regulate ubiquitination, trafficking, and stability of the mGlu7 receptor
Figures
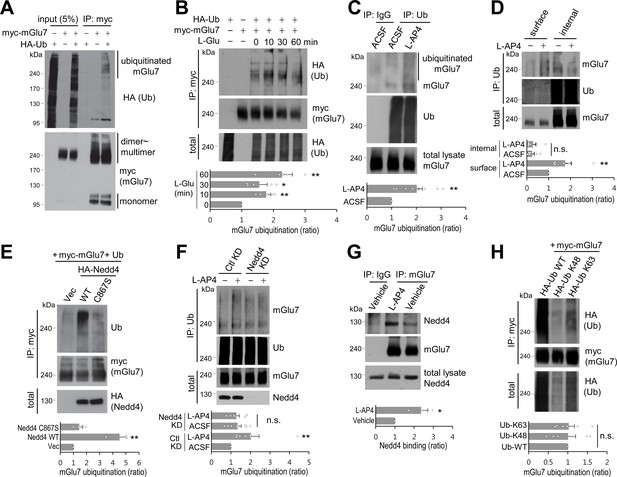
mGlu7 is ubiquitinated by agonist treatment or Nedd4 expression in heterologous cells and neurons.
(A) mGlu7 is ubiquitinated in HEK 293 T cells. N-terminal c-myc epitope-tagged mGlu7a (myc-mGlu7) was co-transfected with HA epitope-tagged ubiquitin (HA-Ub) in HEK 293 T cells. Cell lysates were immunoprecipitated with anti-myc antibody (9E10) and immunoblotted with anti-HA antibody. Diffuse bands of high molecular weight larger than 200 kDa representing ubiquitinated mGlu7 were detected in the lane that the co-expressed mGlu7 and Ub were loaded in. (B) HA-Ub and myc-mGlu7 were co-transfected in HEK 293 T cells. Agonist L-Glutamate (L-Glu, 1 mM) was added for 10 to 60 min before cell lysis. Bar graph below represents L-Glu-induced ubiquitination levels normalized to the 0 min timepoint sample (means ± SEM; 10 min, 1.77 ± 0.15; 30 min, 1.55 ± 0.25; 60 min, 2.27 ± 0.33; n = 5, *p<0.05, **p<0.01 versus 0 min, one-way ANOVA). (C) Cultured cortical neurons at 14 days in vitro (DIV) were incubated in ACSF (140 mM NaCl, 5 mM KCl, 10 mM HEPES, 10 mM D-glucose, 2 mM CaCl2, 2 mM MgCl2 plus 10 μM MG132 and 52.5 μM leupeptin) for 10 min and then 400 μM L-AP4 was added for 5 min at 37°C. Endogenous mGlu7 ubiquitination in neurons was detected by immunoprecipitation using anti-ubiquitin antibody (FK2) and western blotting using anti-ubiquitin (P4D1) or anti-mGlu7 antibody. Bar graph below represents mean ± SEM (L-AP4, 2.02 ± 0.23; n = 9, **p<0.01, Student’s t-test). (D) After cell surface biotinylation with membrane impermeable Sulfo-NHS-SS-Biotin, cortical neurons were treated with L-AP4 for 5 min. The surface proteins were isolated by Streptavidin-agarose beads overnight at 4°C and eluted by incubating the beads with 50 mM DTT for 30 min at 50°C. The eluted proteins (surface) and unbound lysates (internal) were further immunoprecipitated by anti-ubiquitin antibody (FK2), and western blotted with the indicated antibodies. Bar graph below represents means ± SEM of relative mGlu7 ubiquitination levels normalized to total mGlu7 and are shown as a ratio to the non-stimulated control of the surface fraction (surface, L-AP4, 1.75 ± 0.28; internal, ACSF, 0.25 ± 0.09; internal, L-AP4, 0.28 ± 0.13; n = 6, **p<0.01, one-way ANOVA). (E) Nedd4 E3 ligase regulates agonist-stimulated ubiquitination of mGlu7. HA-tagged Nedd4 WT or C867S mutant was co-expressed with Ub and myc-mGlu7 in HEK 293 T cells. Ubiquitination of mGlu7 was analyzed as above. Bar graph below represents mean ± SEM of Nedd4-induced mGlu7 ubiquitination levels normalized to the vector control (Nedd4 WT, 4.51 ± 0.52; Nedd4 C867S, 1.44 ± 0.27; n = 3, **p<0.01, one-way ANOVA). (F) Cultured cortical neurons were infected with lentiviruses harboring Nedd4 shRNA (Nedd4 KD) or non-related control shRNA (Ctl KD) for 7 days. At DIV 14, L-AP4-induced endogenous mGlu7 ubiquitination was evaluated as in Figure 1C. Bar graph represents means ± SEM (Ctl KD + L-AP4, 2.03 ± 0.41; Nedd4 KD + ACSF, 1.33 ± 0.19; Nedd4 KD + L-AP4, 1.28 ± 0.14; n = 7, **p<0.01, n.s. indicates p>0.05, one-way ANOVA). (G) Analysis of endogenous interaction between Nedd4 E3 ligase and mGlu7. Cortical neurons at DIV 14 were treated with L-AP4 for 5 min, and cell lysates were immunoprecipitated with anti-mGlu7 antibody and western blotting was carried out with the indicated antibodies. Bar graph represents mean ± SEM (L-AP4, 2.36 ± 0.36; n = 3, *p<0.05, Student’s t-test). (H) HA-tagged K48 or K63 Ub was co-transfected with mGlu7 in HEK 293 T cells. The cells were treated with MG132 (10 μM) and leupeptin (52.5 μM) for 5 hr to inhibit the proteasomal and lysosomal degradation of the receptor. The cells were stimulated with L-Glu for 5 min before harvesting. K48- or K63-mediated ubiquitination levels were quantified after normalization to total ubiquitination levels. Bar graph below represents mGlu7 band intensities normalized to Ub-WT (means ± SEM; Ub-K48, 1.01 ± 0.19; Ub-K63, 1.02 ± 0.15; n = 6, n.s. indicates p>0.05, one-way ANOVA).
-
Figure 1—source data 1
Ubiquitination of mGlu7 in heterologous cells and neurons.
- https://doi.org/10.7554/eLife.44502.004
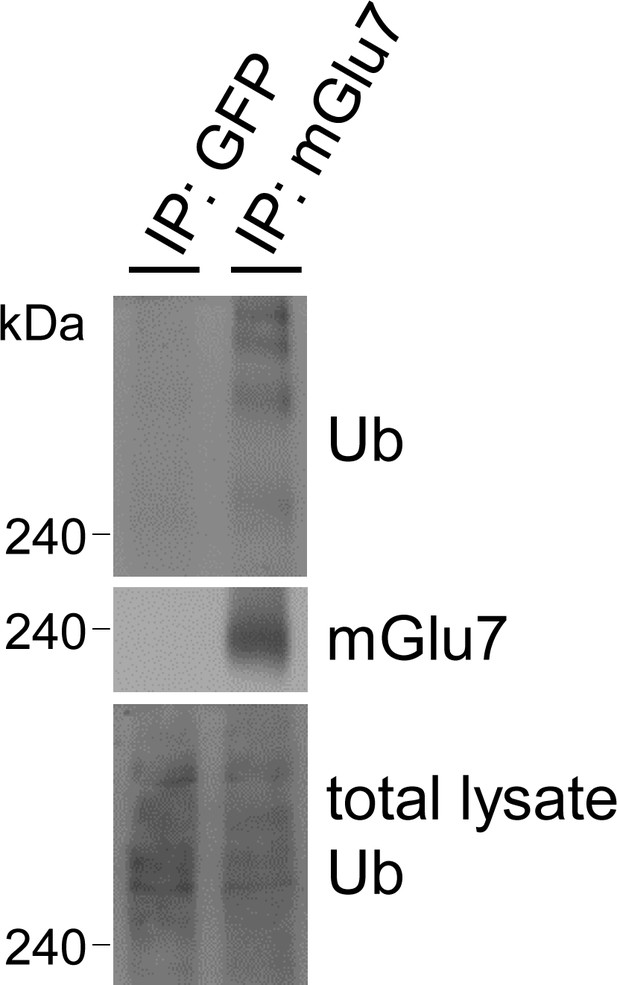
Endogenous mGlu7 is ubiquitinated in cortical neurons.
After cortical neurons were treated with 800 μM L-AP4 for 5 min, the lysates were immunoprecipitated with rabbit anti-mGlu7 antibody. Rabbit anti-GFP antibody was used as a negative control. Immunoprecipitates were separated by SDS-PAGE and analyzed by western blotting using the indicated antibodies.
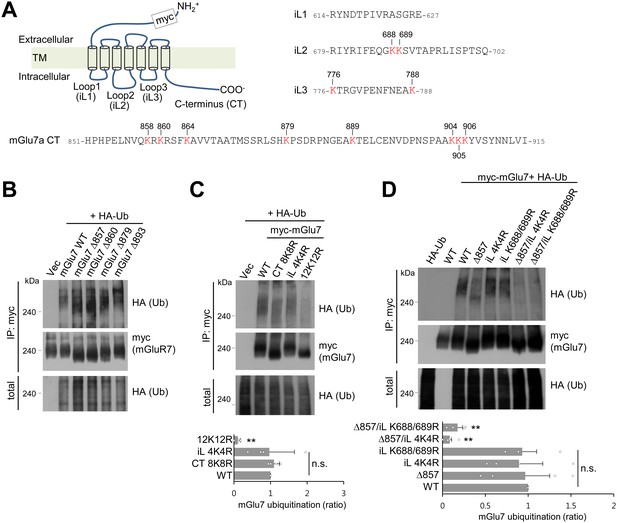
mGlu7 is ubiquitinated at lysine residues in both CT and iL2 domains.
(A) A schematic diagram showing amino acid sequences of mGlu7 intracellular loop regions (iL1‒3) and the cytoplasmic C-terminus tail (CT) of rat mGlu7a. The twelve lysine (K) residues in the CT and iL domains are displayed in red. (B) Myc-mGlu7 WT or sequential deletion mutants at the designated position were co-transfected with HA-Ub in HEK 293 T cells. Cell lysates were immunoprecipitated with anti-myc antibody and immunoblotted with anti-HA antibody. Diffuse bands of high molecular weight larger than 200 kDa represent ubiquitinated mGlu7. (C) Ubiquitination of mGlu7 WT or mutants in which the lysine (K) residues have been substituted to arginine (R) residues. Ubiquitination levels were analyzed as in Figure 1A after 5 min treatment of 1 mM L-Glu. CT 8K8R had all eight lysine residues in the mGlu7 CT mutated to arginines; iL 4K4R, all four lysine residues in the mGlu7 iL domains to arginines; 12K12R, all twelve lysine residues in the mGlu7 CT and iL domains to arginines. CT, C-terminal tail; iL, intracellular loop. Bar graph below represents mean ± SEM showing quantification of ubiquitination levels in the mutants normalized to WT (CT 8K8R, 1.09 ± 0.08; iL 4K4R, 0.97 ± 0.34; 12K12R, 0.11 ± 0.04; n = 4, **p<0.01, n.s. indicates p>0.05, one-way ANOVA). (D) Ubiquitination of mGlu7 in the intracellular loops in HEK 293 T cells. mGlu7 Δ857 does not harbor any lysine residues in the CT due to the stop codon at amino acid position 857. mGlu7 iL K688/689R represents two lysine residues in the iL2 are mutated to arginines. mGlu7 Δ857/iL 4K4R or mGlu7 Δ857/iL K688/689R are mutants that combine Δ857 and iL 4K4R or iL K688/689R mutations, respectively. Bar graph below represents mean ± SEM of band intensities normalized to WT (Δ857, 0.97 ± 0.29; iL 4K4R, 0.89 ± 0.28; iL K688/689R, 0.93 ± 0.17; Δ857/iL 4K4R, 0.08 ± 0.02; Δ857/iL K688/689R, 0.17 ± 0.06; n = 4, **p<0.01, n.s. indicates p>0.05, one-way ANOVA).
-
Figure 2—source data 1
Mapping lysine residues of mGlu7 ubiquitination.
- https://doi.org/10.7554/eLife.44502.006
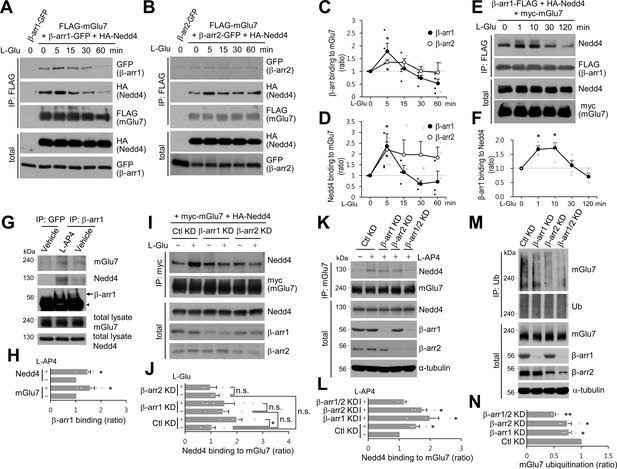
β-arrestin 1 (β-arr1), β-arrestin 2 (β-arr2), and Nedd4 are recruited to mGlu7 by agonist stimulation within 5 min.
(A–F) Time-course interaction among mGlu7, Nedd4, and β-arrestins. HEK 293 T cells were co-transfected with FLAG-mGlu7, HA-Nedd4, and β-arrestin 1-GFP (A) or β-arrestin 2-GFP (B). In panel E, myc-mGlu7, HA-Nedd4, and β-arrestin 1-FLAG were utilized. L-Glu (1 mM) was administered for the indicated time before cell lysis. Cell lysates were immunoprecipitated with anti-FLAG antibody and western blotting was carried out with the indicated antibodies. Time-course binding of β-arrestins to mGlu7 (C), Nedd4 to mGlu7 (D), β-arrestin 1 to Nedd4 (F) were quantified and presented as mean ± SEM (C, β-arr1, 5 min, 1.79 ± 0.31; 15 min, 1.11 ± 0.23; 30 min, 0.73 ± 0.12; 60 min, 0.52 ± 0.17; β-arr2, 5 min, 1.36 ± 0.10; 15 min, 1.36 ± 0.15; 30 min, 1.01 ± 0.18; 60 min, 0.96 ± 0.38; D, β-arr1, 5 min, 2.36 ± 0.57; 15 min, 1.16 ± 0.31; 30 min, 0.61 ± 0.14; 60 min, 0.72 ± 0.49; β-arr2, 5 min, 2.20 ± 0.36; 15 min, 1.97 ± 0.68; 30 min, 1.96 ± 0.62; 60 min, 1.83 ± 0.49; F, β-arr1, 1 min, 1.67 ± 0.12; 10 min, 1.72 ± 0.19; 30 min, 1.11 ± 0.17; 120 min, 0.71 ± 0.13; n = 3‒4, *p<0.05 versus 0 min, Student’s t-test). (G) Endogenous β-arrestin 1, Nedd4, and mGlu7 are present in the same complex. After cortical neurons were treated with 400 μM L-AP4 for 5 min, the lysates were immunoprecipitated with anti-β-arrestin 1 antibody or anti-GFP antibody as a control. Arrow and arrowhead indicate β-arrestin 1 and immunoglobulin heavy chain (IgH), respectively. (H) Bar graph represents mean ± SEM showing quantification of mGlu7 or Nedd4 binding levels to β-arrestin 1 (mGlu7 L-AP4, 1.56 ± 0.19; Nedd4 L-AP4, 1.46 ± 0.13; n = 5, *p<0.05 versus vehicle, Student’s t-test). (I) myc-mGlu7 and HA-Nedd4 were co-transfected either with pSuper β-arrestin 1, 2 shRNA or pSuper control shRNA in HEK 293 T cells. Three days after transfection, 1 mM L-Glu was treated for 5 min and cell lysates were immunoprecipitated using anti-myc antibody. Immunoprecipitates were resolved by SDS-PAGE and analyzed by western blotting using the indicated antibodies. (J) Bar graph represents mean ± SEM showing Nedd4 binding to mGlu7 levels normalized to untreated Ctl KD (Ctl KD + L-Glu, 1.98 ± 0.22; β-arr1 KD, 1.46 ± 0.23; β-arr1 KD + L-Glu, 1.51 ± 0.39; β-arr2 KD, 1.19 ± 0.13; β-arr2 KD + L-Glu, 0.97 ± 0.25; n = 3‒5, *p<0.05, n.s. indicates p>0.05, one-way ANOVA). (K) β-arrestins are involved in agonist-induced Nedd4 recruitment to mGlu7 in neurons. Cultured cortical neurons were infected with lentiviruses harboring β-arrestin 1 or 2 shRNA (β-arr1 or 2 KD), or non-related target shRNA (Ctl KD) for 7 days. Both β-arrestin 1 and 2 shRNA were co-infected for β-arr1/2 KD lane. After treatment with 400 μM L-AP4 for 5 min, the lysates were immunoprecipitated with anti-mGlu7 antibody and endogenous Nedd4 binding was analyzed by western blotting. (L) Bar graph represents mean ± SEM showing quantification of Nedd4 binding levels to mGlu7 (Ctl KD + L-AP4, 1.54 ± 0.11; β-arr1 KD + L-AP4, 1.99 ± 0.29; β-arr2 KD + L-AP4, 1.73 ± 0.16; β-arr1/2 KD, 1.15 ± 0.07; n = 4, *p<0.05 versus vehicle in Ctl KD, Student’s t-test). (M) β-arrestins mediate agonist-induced ubiquitination of mGlu7 in neurons. Cultured cortical neurons were infected with β-arrestins shRNA lentiviruses and then incubated with L-AP4 (400 μM) as panel K. Cell lysates were immunoprecipitated with anti-ubiquitin antibody (FK2) and western blotting was performed with anti-mGlu7 antibody. (N) Bar graph represents mean ± SEM showing mGlu7 ubiquitination levels normalized to Ctl KD (β-arr1 KD, 0.77 ± 0.06; β-arr2 KD, 0.74 ± 0.08; β-arr1/2 KD, 0.50 ± 0.03; n = 4, **p<0.01, *p<0.05, Student’s t-test).
-
Figure 3—source data 1
Binding analysis of mGlu7, Nedd4, and beta-arrestins complex.
- https://doi.org/10.7554/eLife.44502.015
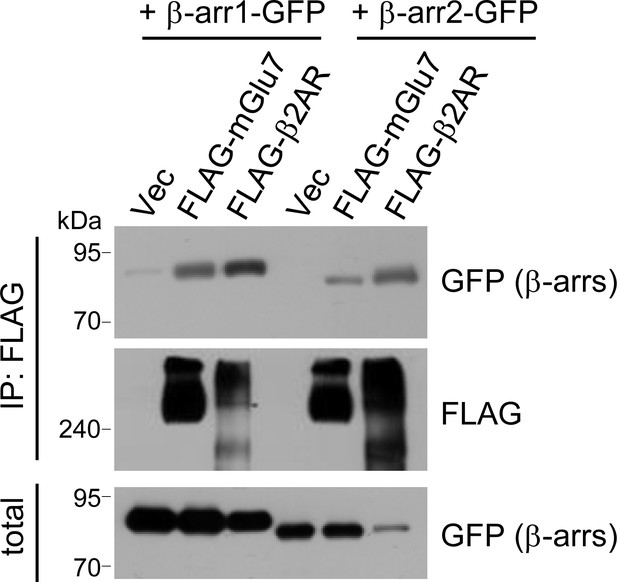
Binding of β-arrestins to mGlu7.
GFP-tagged β-arrestin 1 or 2 was co-transfected with either FLAG-tagged mGlu7 or FLAG-tagged β2AR in HEK 293 T cells. The lysates were immunoprecipitated with anti-FLAG antibody and western blotting was carried out with the indicated antibodies.
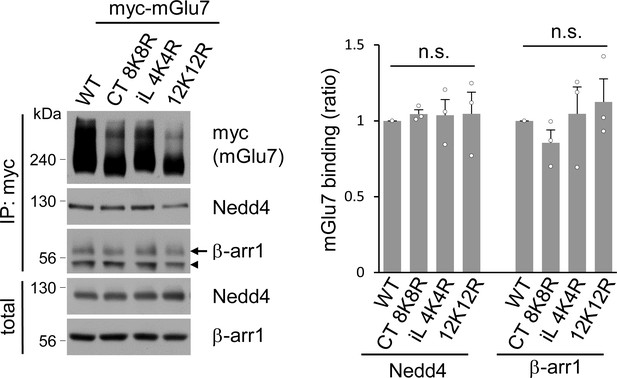
The binding affinity of Nedd4 and β-arrestin 1 to mGlu7 is not altered by ubiquitination site mutations of mGlu7.
mGlu7 WT, iL 4K4R, Ct 8K8R, or 12K12R was expressed on cultured cortical neurons via lentivirus-mediated transduction. After treatment with 400 μM L-AP4 for 5 min, co-immunoprecipitation assay was carried out using anti-myc antibody. Arrow and arrowhead indicate β-arrestin 1 and immunoglobulin heavy chain (IgH), respectively. Bar graph in the right panel represents mean ± SEM showing quantification of Nedd4 or β-arrestin 1 binding levels in the mutants normalized to WT (Nedd4, CT 8K8R, 1.05 ± 0.03; iL 4K4R, 1.04 ± 0.10; 12K12R, 1.05 ± 0.14; β-arr1, CT 8K8R, 0.85 ± 0.09; iL 4K4R, 1.05 ± 0.18; 12K12R, 1.13 ± 0.15; n = 3, n.s. indicates p>0.05, Student’s t-test).
-
Figure 3—figure supplement 2—source data 1
Binding affinity of Nedd4 and beta-arrestin 1 to mGlu7 ubiquitination site mutants.
- https://doi.org/10.7554/eLife.44502.010
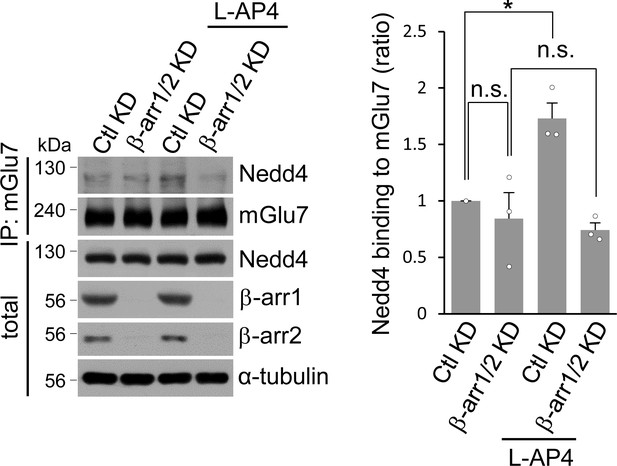
Co-immunoprecipitation assay to supplement Figures 3K and L.
Bar graph represents mean ± SEM showing quantification of binding affinity between Nedd4 and mGlu7 normalized to Ctl KD (β-arr1/2 KD, 0.84 ± 0.23; Ctl KD + L-AP4, 1.73 ± 0.14; β-arr1/2 KD + L-AP4, 0.74 ± 0.06; n = 3, *p<0.05 versus vehicle in Ctl KD, n.s. indicates p>0.05, Student’s t-test).
-
Figure 3—figure supplement 3—source data 1
Binding of Nedd4 to mGlu7 by beta-arrestins in neurons.
- https://doi.org/10.7554/eLife.44502.012

Ubiquitination assay to supplement Figures 3M and N.
Bar graph represents mean ± SEM showing mGlu7 ubiquitination levels normalized to Ctl KD (β-arr1/2 KD, 1.15 ± 0.09; Ctl KD + L-AP4, 1.42 ± 0.07; β-arr1/2 KD + L-AP4, 1.06 ± 0.09; n = 3, *p<0.05 versus vehicle in Ctl KD, n.s. indicates p>0.05, Student’s t-test).
-
Figure 3—figure supplement 4—source data 1
Ubiquitination of mGlu7 by beta-arrestins in neurons.
- https://doi.org/10.7554/eLife.44502.014

Mapping of binding domains between mGlu7 and Nedd4 or β-arrestins.
(A) GST pull-down experiments to identify the interaction domains of mGlu7 with β-arrestin 1 or Nedd4. Total rat brain extract was subjected to a pull-down assay using immobilized GST fusion proteins containing the mGlu7 iL 1–3 domains, mGlu7a CT, or mGlu7b CT. After washing, bound proteins were analyzed by SDS-PAGE and western blotting using the indicated antibodies. The amount of GST fusion protein as bait was verified by Coomassie Brilliant Blue (CBB) staining. (B) Direct binding assay between mGlu7 and Nedd4 or β-arrestins. Recombinant His-tagged Nedd4, β-arrestin 1, or β-arrestin 2 proteins were purified using Ni-NTA resin. The purified proteins were incubated at 4°C for 2 hr with immobilized GST-mGlu7 iL or CT domains. After washing, bound proteins were analyzed by SDS-PAGE and western blotting using the indicated antibodies. (C) Mapping of binding domain of Nedd4 to mGlu7. Schematic diagrams of Nedd4 where each domain has been deleted are shown. ΔC2, amino acids (aa) 1‒151; ΔC2-WW1/2, aa 1‒380; ΔWW3, aa 381‒454; ΔWW4, aa 455‒506; ΔHECT, aa 507‒900; Δ denotes deletion. (D) mGlu7 binds to the HECT domain of Nedd4. myc-mGlu7 was co-transfected with the indicated FLAG-Nedd4 domain deletion constructs in HEK 293 T cells. Immunoprecipitation was carried out with anti-myc antibody, and bound proteins were detected with anti-FLAG antibody.
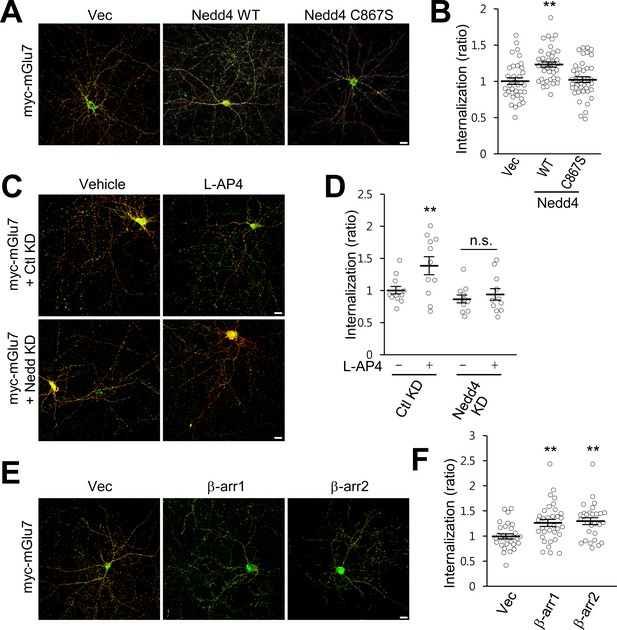
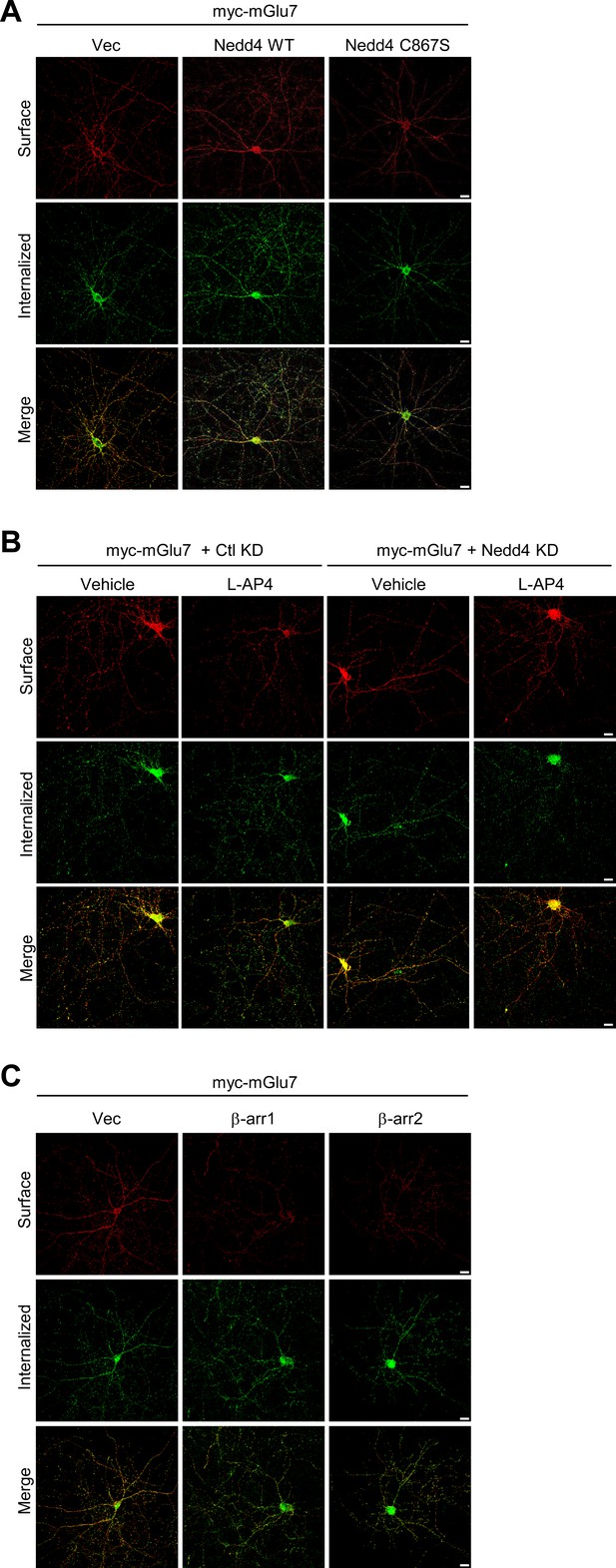
β-arrestins and Nedd4 regulate endocytosis of mGlu7 in neurons.
(A) The endocytosis of mGlu7 was analyzed by an antibody uptake internalization assay. myc-mGlu7 was co-transfected with Nedd4 WT, C867S mutant, or vector control (Vec) in cultured hippocampal neurons. Two days after transfection, neurons were labeled with anti-myc antibody for 10 min, and returned to conditioned media for 15 min at 37°C. Neurons were fixed and incubated with Alexa Fluor 568-conjugated secondary antibody (red) to label surface-expressed receptors before permeabilization. After permeabilization with 0.25% Trion X-100 for 5 min, neurons were then incubated with Alexa Fluor 488-conjugated secondary antibody (green) to label the internalized receptors. Merged images are presented in which the red signal represents the surface mGlu7 and the green signal represents the internalized mGlu7. Scale bar, 20 μm. (B) Summary histograms quantifying the internalized mGlu7 from panel A are present as the ratio of the internalized population compared with total (surface + internalized) population measured using Metamorph software. Scatter plots show mean ± SEM (Vec, 1.00 ± 0.04; Nedd4 WT, 1.23 ± 0.04; Nedd4 C867S, 1.02 ± 0.04; n > 35, **p<0.01, one-way ANOVA). (C) myc-mGlu7 was co-transfected with pSuper-Ctl shRNA (Ctl KD) or pSuper-Nedd4 shRNA (Nedd4 KD) in cultured hippocampal neurons. Internalization of mGlu7 was analyzed in the absence or presence of 400 μM L-AP4 for 15 min at 37°C. (D) Summary histograms quantifying the internalized mGlu7 from panel C. Scatter plots show mean ± SEM (Vec, 1.00 ± 0.06; Vec + L-AP4, 1.39 ± 0.14; Nedd4 KD, 0.86 ± 0.06; Nedd4 KD + L-AP4, 0.94 ± 0.09; n > 10, *p<0.05, n.s. indicates p>0.05, one-way ANOVA). (E) myc-mGlu7 and β-arrestin 1 or 2 were co-expressed and the internalized mGlu7 was analyzed as above. (F) Summary histograms quantifying the internalized mGlu7 from panel E. Scatter plots show mean ± SEM (Vec, 1.00 ± 0.05; β-arr1, 1.26 ± 0.07; β-arr2, 1.30 ± 0.07; n > 30, **p<0.01, one-way ANOVA).
-
Figure 5—source data 1
Endocytosis of mGlu7 by beta-arrestins and Nedd4.
- https://doi.org/10.7554/eLife.44502.020

Ubiquitination is required for endocytosis of endogenous mGlu7.
Cortical neurons were treated with 0.1 μM MLN7243 for 18 hr to inhibit ubiquitination. After treatment with 800 μM L-AP4 for 15 min, surface biotinylation assay was performed. Treatment of MLN7243 in cortical neurons resulted in increased surface expression and decreased ubiquitination of endogenous mGlu7.
Separated images that supplement the merged images shown in Figure 5.
Surface-expressed and internalized mGlu7 signals in Figures 5A, C and E were separated, and individual images were presented in A, B, and C, respectively.
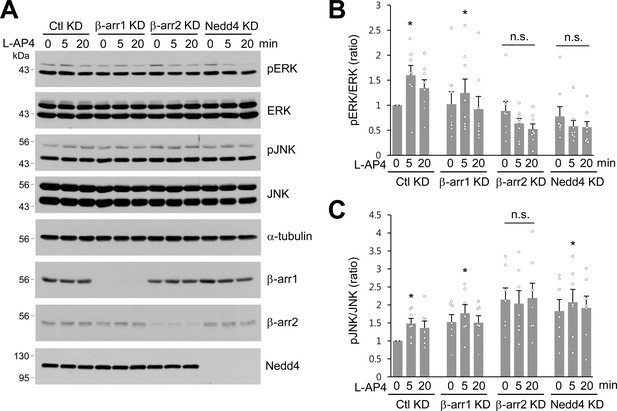
β-arrestins and Nedd4 regulate ERK and JNK signaling of mGlu7 in neurons.
(A) Cultured hippocampal neurons were infected with control (Ctl KD), β-arrestin 1 (β-arr1 KD), β-arrestin 2 (β-arr2 KD), or Nedd4 KD shRNA lentiviruses for 7 days. At DIV 14, neurons were treated with 400 μM L-AP4 for 0, 5, 20 min, and neuronal lysates were separated by SDS-PAGE and probed using the indicated antibodies. (B) Bar graph represents mean ± SEM of pERK band intensities normalized to the control lane (Ctl KD, 5 min, 1.60 ± 0.20; 20 min, 1.34 ± 0.17; β-arr1 KD, 0 min, 1.02 ± 0.25; 5 min, 1.25 ± 0.28; 20 min, 0.92 ± 0.26; β-arr2 KD, 0 min, 0.89 ± 0.19; 5 min, 0.64 ± 0.10; 20 min, 0.53 ± 0.10; Nedd4 KD, 0 min, 0.78 ± 0.20; 5 min, 0.58 ± 0.12; 20 min, 0.56 ± 0.11; n = 8, *p<0.05, n.s. indicates p>0.05, Student’s t-test). (C) Bar graph represents mean ± SEM of pJNK band intensities normalized to the control lane (Ctl KD 5 min, 1.48 ± 0.14; 20 min, 1.36 ± 0.19; β-arr1 KD 0 min, 1.52 ± 0.21; 5 min, 1.77 ± 0.24; 20 min, 1.50 ± 0.19; β-arr2 KD 0 min, 2.15 ± 0.33; 5 min, 2.03 ± 0.36; 20 min, 2.19 ± 0.41; Nedd4 KD 0 min, 1.83 ± 0.32; 5 min, 2.07 ± 0.36; 20 min, 1.92 ± 0.33; n = 7, *p<0.05, n.s. indicates p>0.05, Student’s t-test).
-
Figure 6—source data 1
ERK and JNK signaling of mGlu7 by beta-arrestins and Nedd4.
- https://doi.org/10.7554/eLife.44502.022
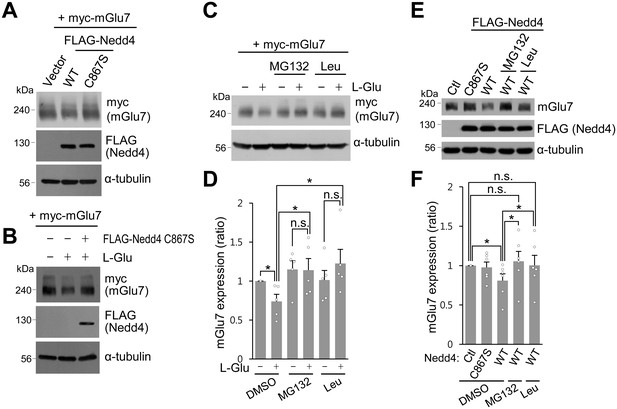
Nedd4-mediated or agonist-stimulated mGlu7 degradation occurs via both the proteasomal and lysosomal degradation pathways.
(A) myc-mGlu7 and FLAG-Nedd4 WT or C867S were co-transfected in HEK 293 T cells. Thirty-six hours after transfection, cell lysates were analyzed by western blotting using the indicated antibodies. (B) Following transfection, cells were starved overnight in serum-free DMEM culture medium and then treated with 1 mM L-Glu for 1 hr. Cell lysates were analyzed by western blotting using the indicated antibodies. (C) HEK 293 T cells transiently expressing myc-tagged mGlu7 was incubated with 100 μM MG132 or 52.5 μM leupeptin (Leu) for 2 hr and then L-Glu was added for 1 hr. Cell lysates were analyzed by SDS-PAGE and western blotting using the indicated antibodies. (D) Summary histograms quantifying mGlu7 expression in panel C. Bar graph represents mean ± SEM of band intensities normalized to control lane (DMSO + L-Glu, 0.74 ± 0.09; MG132, 1.15 ± 0.11; MG132 + L-Glu, 1.14 ± 0.15; Leu, 1.01 ± 0.12; Leu + L-Glu, 1.23 ± 0.18; n = 5, *p<0.05, n.s. indicates p>0.05, Student’s t-test). (E) FLAG-tagged Nedd4 WT or Nedd4 C867S was expressed on cultured cortical neurons via lentiviruses. After treatment with 0.5 μg/ml cycloheximide for 21 hr, the neurons were incubated with 100 μM MG132 or 52.5 μM Leu for 4 hr and then 400 μM L-AP4 was added for 1 hr. Cell lysates were analyzed by western blotting using the indicated antibodies. (F) Summary histograms quantifying mGlu7 expression in panel E. Bar graph represents mean ± SEM of band intensities normalized to control lane (Nedd4 C867S, 0.98 ± 0.07; Nedd4 WT, 0.81 ± 0.09; Nedd4 WT + MG132, 1.06 ± 0.13; Nedd4 WT + Leu, 1.00 ± 0.13; n = 5, *p<0.05, n.s. indicates p>0.05, Student’s t-test).
-
Figure 7—source data 1
Degradation pathways of ubiquitinated mGlu7.
- https://doi.org/10.7554/eLife.44502.024
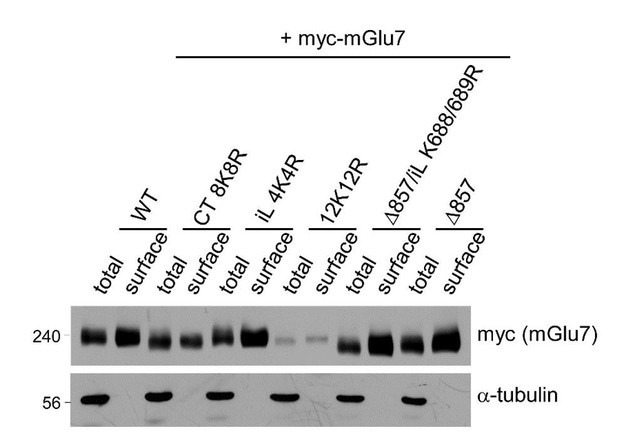
Surface expression levels of mGlu7 ubiquitination site mutants by cell surface biotinylation assay. mGlu7 WT, iL 4K4R, Ct 8K8R, 12K12R, Δ857, or Δ857/iL K688/689R mutants were expressed in cultured cortical neurons for 7 days via lentivirus-mediated transduction.
At DIV14, after cell surface biotinylation with membrane impermeable Sulfo-NHS-LC-Biotin, surface proteins were isolated by Streptavidin-agarose beads. Western blotting was performed with the indicated antibodies.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pRK5-myc-mGlu7 WT | Dr. Katherine W. Roche (NIH) | ||
| Recombinant DNA reagent | pRK5-FLAG-mGlu7 WT | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ857 | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ860 | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ879 | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ893 | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 CT 8K8R | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 iL 4K4R | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 12K12R | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 iL K688/689R | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ857/iL 4K4R | This study | ||
| Recombinant DNA reagent | pRK5-myc-mGlu7 Δ857/IL K688/689R | This study | ||
| Recombinant DNA reagent | FLAG-ß2AR | Addgene | RRID:Addgene_14697 | |
| Recombinant DNA reagent | pRK5-HA-Ub WT | Addgene | RRID:Addgene_17608 | |
| Recombinant DNA reagent | pRK5-HA-Ub K48 | Addgene | RRID:Addgene_17605 | |
| Recombinant DNA reagent | pRK5-HA-Ub K63 | Addgene | RRID:Addgene_17606 | |
| Recombinant DNA reagent | pGFP-N3-ß-arrestin1-GFP | Dr. Katherine W. Roche (NIH) | ||
| Recombinant DNA reagent | pGFP-N3-ß-arrestin2-GFP | Dr. Katherine W. Roche (NIH) | ||
| Recombinant DNA reagent | pcDNA3-ß-arrestin1-FLAG | Addgene | RRID:Addgene_14687 | |
| Recombinant DNA reagent | pCI-HA-Nedd4 WT | Addgene | RRID:Addgene_27002 | |
| Recombinant DNA reagent | pCI-HA-Nedd4 C867S | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 WT | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 C867S | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 ΔC2 | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 ΔC2-WW1/2 | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 ΔWW3 | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 ΔWW4 | This study | ||
| Recombinant DNA reagent | FLAG-Nedd4 ΔHECT | This study | ||
| Recombinant DNA reagent | pSuper-rat Nedd4 KD | This study | ||
| Recombinant DNA reagent | pSuper-human ß-arrestin1 KD | This study | ||
| Recombinant DNA reagent | pSuper-human ß-arrestin2 KD | This study | ||
| Recombinant DNA reagent | pSuper-rat ß-arrestin1 KD | This study | ||
| Recombinant DNA reagent | pSuper-rat ß-arrestin2 KD | This study | ||
| Recombinant DNA reagent | pSuper-non-related target KD | This study | ||
| Recombinant DNA reagent | FHUGW-rat ß-arrestin1 KD | This study | ||
| Recombinant DNA reagent | FHUGW-rat ß-arrestin2 KD | This study | ||
| Recombinant DNA reagent | FHUGW-non-related target KD | This study | ||
| Recombinant DNA reagent | FHUGW-myc-mGlu7 WT | This study | ||
| Recombinant DNA reagent | FHUGW-myc-mGlu7 CT 8K8R | This study | ||
| Recombinant DNA reagent | FHUGW-myc-mGlu7 iL 4K4R | This study | ||
| Recombinant DNA reagent | FHUGW-myc-mGlu7 12K12R | This study | ||
| Recombinant DNA reagent | FHUGW-FLAG-Nedd4 WT | This study | ||
| Recombinant DNA reagent | FHUGW-FLAG-Nedd4 C867S | This study | ||
| Recombinant DNA reagent | pGEX-4T-1-mGlu7 iL1 | This study | ||
| Recombinant DNA reagent | pGEX-4T-1-mGlu7 iL2 | This study | ||
| Recombinant DNA reagent | pGEX-4T-1-mGlu7 iL3 | This study | ||
| Recombinant DNA reagent | pGEX-4T-1-mGlu7a CT | This study | ||
| Recombinant DNA reagent | pGEX-4T-1-mGlu7b CT | This study | ||
| Recombinant DNA reagent | pET28a-Nedd4 | This study | ||
| Recombinant DNA reagent | pET28b-Barr1 | This study | ||
| Recombinant DNA reagent | pET28b-Barr2 | This study | ||
| Antibody | HA (clone 16B12) | BioLegend | cat# 901501 RRID:AB_2565006 | 1:1000, western blotting (WB) |
| Antibody | c-myc (clone 9E10) | SIGMA | cat# M5546 RRID:AB_260581 | 1:1000, WB; 1:500, immunofluorescence staining (IF) |
| Antibody | FLAG (clone M2) | SIGMA | cat# F1804 RRID:AB_262044 | 1:1000, WB |
| Antibody | FLAG | SIGMA | cat# F7425 RRID:AB_439687 | 1:1000, WB |
| Antibody | mGlu7a | EMD Millipore | cat# 07–239 RRID:AB_310459 | 1:2000, WB |
| Antibody | Ub (clone FK2) | EMD Millipore | cat# 04–263 RRID:AB_612093 | 1:1000, WB |
| Antibody | Ub (clone P4D1) | Santa Cruz Biotechnology | cat# sc-8017 RRID:AB_2762364 | 1:1000, WB |
| Antibody | Nedd4 | EMD Millipore | cat# 07–049 RRID:AB_310351 | 1:10000, WB |
| Antibody | Nedd4 | R&D SYSTEMS | cat# MAB6218 RRID:AB_10920762 | 1:1000, WB |
| Antibody | Beta-arrestin 1 (clone E274) | Abcam | cat# ab32099 RRID:AB_722896 | 1:1000, WB |
| Antibody | Beta-arrestin 2 (clone C16D9) | Cell Signaling Technology | cat# 3857 RRID:AB_2258681 | 1:1000, WB |
| Antibody | GFP (clone B2) | Santa Cruz Biotechnology | cat# sc-9996 RRID:AB_627695 | 1:500, WB |
| Antibody | GFP | Thermo Fischer Scientific | cat# A11122 RRID:AB_221569 | 1:2000, WB |
| Antibody | ɑ-tubulin | SIGMA | cat# T6199 RRID:AB_477583 | 1:5000, WB |
| Antibody | PP1γ1 | EMD Millipore | cat# 07–1218 RRID:AB_1977432 | 1:500, WB |
| Antibody | phospho-ERK | Santa Cruz Biotechnology | cat# sc-7383 RRID:AB_627545 | 1:1000, WB |
| Antibody | ERK | Santa Cruz Biotechnology | cat# sc-94 RRID:AB_2140110 | 1:2000, WB |
| Antibody | phospho-JNK | Cell Signaling Technology | cat# 4671 RRID:AB_331338 | 1:1000, WB |
| Antibody | JNK | Cell Signaling Technology | cat# 9252 RRID:AB_2250373 | 1:1000, WB |
| Antibody | Goat anti-Mouse IgG (H+L) antibody, Alexa Fluor 568 | Thermo Fischer Scientific | cat# A11031 RRID:AB_144696 | 1:500, IF |
| Antibody | Goat anti-Mouse IgG (H+L) antibody, Alexa Fluor 488 | Thermo Fischer Scientific | cat# A11029 RRID:AB_138404 | 1:500, IF |
| Chemical compound | Normal goat serum blocking solution | VECTOR LABORATORIES | cat# S-1000 RRID:AB_2336615 | |
| Chemical compound | Paraformaldehyde | SIGMA | cat# 158127 | |
| Chemical compound | ProLong Antifade Kit | Thermo Fischer Scientific | cat# P7481 | |
| Chemical compound | L-Glutamic acid | TOCRIS | cat# 0218 | |
| Chemical compound | L-AP4 | TOCRIS | cat# 0103 | |
| Chemical compound | N-Ethylmaleimide (NEM) | SIGMA | cat# 04259 | |
| Chemical compound | MG132 | TOCRIS | cat# 1748 | |
| Chemical compound | Leupeptin hemisulfate | TOCRIS | cat# 1167 | |
| Chemical compound | Cycloheximide | SIGMA | cat# C7698 | |
| Chemical compound | MLN7243 | CHEMIETEK | cat# CT-M7243 | |
| Chemical compound | Protein G Sepharose 4 Fast Flow | GE Healthcare | cat# 17061801 | |
| Chemical compound | Protein A Sepharose, from Staphylococcus aureus | SIGMA | cat# P3391 | |
| Chemical compound | Glutathione Sepharose 4B GST-tagged resin | GE Healthcare | cat# 17075601 | |
| Chemical compound | EZ-Link Sulfo-NHS-SS-Biotin | Thermo Fischer Scientific | cat# 21331 | |
| Chemical compound | Pierce Streptavidin Agarose | Thermo Fischer Scientific | cat# 20347 | |
| Chemical compound | shRNA to rat Nedd4 | Schwarz et al., 2010 | Oligonucleotides | |
| Chemical compound | shRNA to rat beta-arrestin 1 | Simard et al., 2013 | Oligonucleotides | |
| Chemical compound | shRNA to rat beta-arrestin 2 | Molteni et al., 2009 | Oligonucleotides | |
| Chemical compound | shRNA to human beta-arrestin 1 | Shenoy et al., 2008 | Oligonucleotides | |
| Chemical compound | shRNA to human beta-arrestin 2 | Skånland et al., 2009 | Oligonucleotides | |
| Chemical compound | shRNA to non-related target | GenScript | Oligonucleotides | |
| Cell line (human) | HEK293T | ATCC | cat# CRL-3216 RRID:CVCL_0063 | |
| Rat (Rattus norvegicus) | Primary cultured neurons | ORIENT BIO | RGD Cat# 734476 RRID:RGD_734476) | SD rat |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44502.025





