The interplay at the replisome mitigates the impact of oxidative damage on the genetic integrity of hyperthermophilic Archaea
Figures
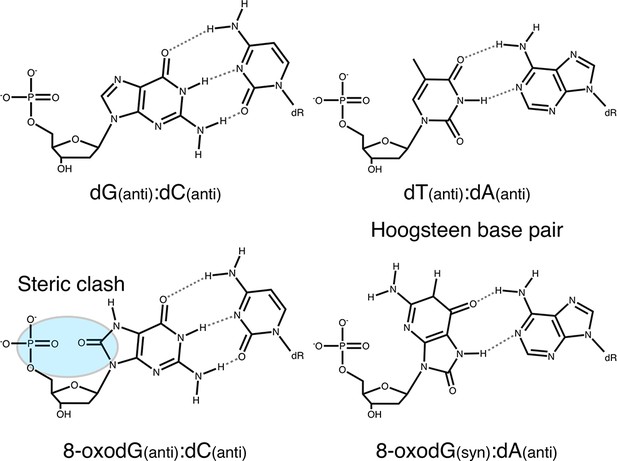
Base pairing of 8-oxoguanosine.
https://doi.org/10.7554/eLife.45320.002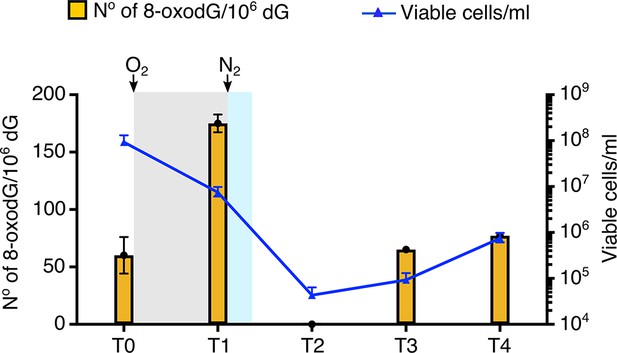
Effect of oxygenation on the viability and the rate of 8-oxodG/106 dG into the genome of P. abyssi.
Oxidative stress is applied to P. abyssi growing cells in a batch mode culture during 5 min. Steady-state level of 8-oxodG per 106 dG and viability are estimated at different times. T0, control before oxidative stress. T1, after the 5 min oxidative stress. T2, 40 min after the 5 min oxidative stress. T3, 140 min after the 5 min oxidative stress. T4, 320 min after the 5 min oxidative stress. Steady-state level of 8-oxodG is calculated from 10 µg of genomic DNA by HPLC-UV-EC as described in the methods. Errors bars indicated analytical duplicates. To enumerate viable cells, most-probable-number (MPN) assays were performed as previously published (Blodgett, 2006) (Oblinger and Koburger, 1975) Survival (cells/ml) is based on a three-tube MNP dilution assay. Upper and lower error bars are shown. Gray and blue shaded indicates the presence of dissolved oxygen in the medium culture. White background corresponds to strict anaerobia. Sparging with oxygen or nitrogen is shown with an arrow. Raw data for each graph are provided in Figure 1—source data 1.
-
Figure 1—source data 1
Quantification of the effect of oxygenation on the viability of P. abyssi cells and the rate rate of 8-oxodG/106 dG in the genome.
- https://doi.org/10.7554/eLife.45320.006
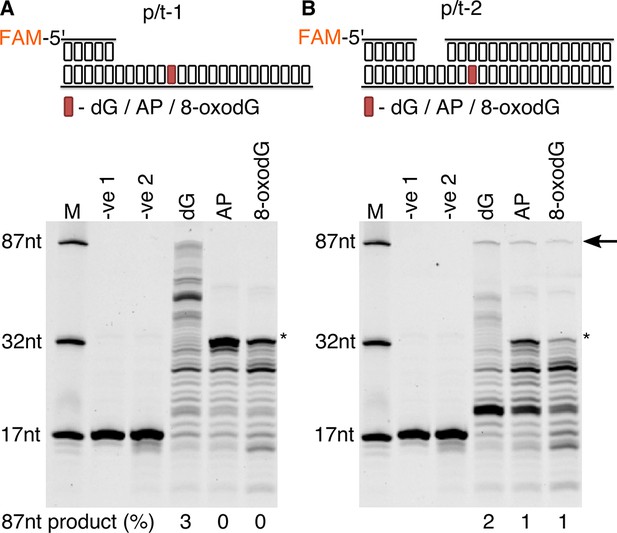
Primer extension and replication bypass of template strand 8-oxodG by P. abyssi cell extracts.
Primer-template extension carried out for 60 min on p/t-1 containing either dG/AP/8-oxodG (A) or p/t-2 containing either dG/AP/8-oxodG (B). In both instances –ve one indicates sample lacking P. abyssi cell extract and –ve two indicates sample containing cell extracts but lacking MgCl2 and dNTPs. An arrow is used to indicate the position of full length extension products (87nt in length), with * used to highlight the approximate location of the damaged base. Shown above each gel is a representative cartoon indicating the structure of primer-templates and the relative position (+33) of the dG/AP/8-oxodG within both DNA primer-template (highlighted in red).
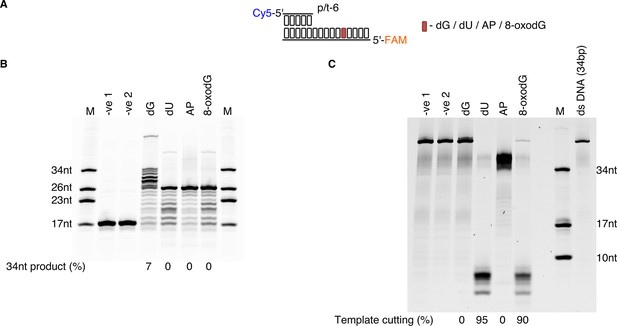
Primer extension and endonuclease activity on 8-oxodG by P. abyssi cell extracts.
(A) Diagrammatic representation of the Cy5 and FAM dual labelled primer-template substrate. (B) Primer extension carried out for 60 min on p/t-6 containing either dG, dU, AP, 8-oxodG with the gel imaged at 633 nm to observe the Cy5 labelled primer. (C) Gel imaged at 532 nm to observe the effects of PabCE on the FAM labelled template strand. Unlabelled ssDNA, complementary to the FAM labelled template, was added to ensure the Cy5 primer DNA product was prevented from re-annealing and visualised as ssDNA as described in the methods. Unfortunately this had the consequence of stabilising any full length FAM labelled template as dsDNA even under denaturing conditions as shown by the presence of the 34 bp dsDNA control. Only FAM labelled DNA that was cut and <10 nt in length migrated as single stranded DNA. For B and C the length of the ladder oligonucleotides in lane M is indicated to the left and right respectively.
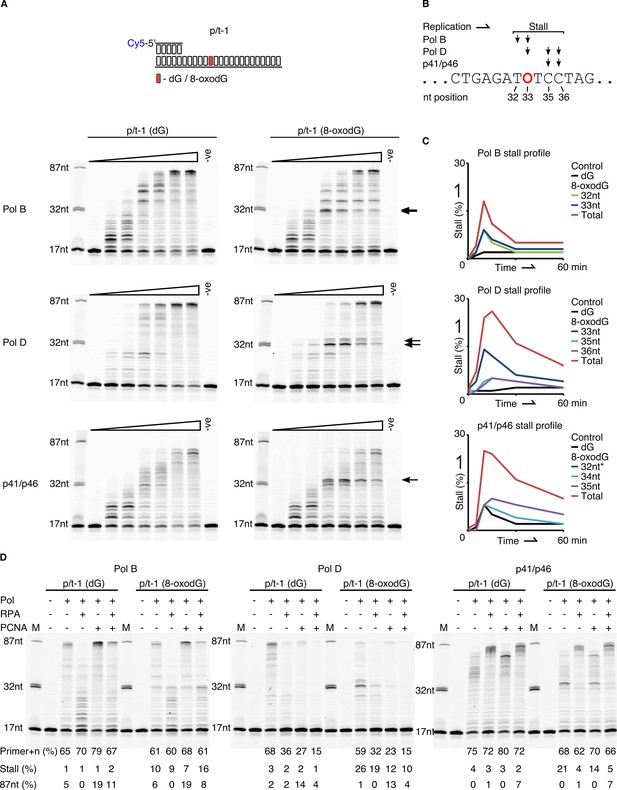
Primer extension and replication bypass of template strand 8-oxodG by P. abyssi replicative proteins.
(A) Primer-template extension reactions performed on p/t-1 containing either dG or 8-oxodG by PolB, PolD and the p41/p46 complex. The triangle above each gel image indicates time course of primer extension (time points taken at 0, 0.5, 1, 5, 10, 30 and 60 min), the arrows to the right of each gel indicates the location of 8-oxodG induced stalling. –ve samples were incubated for 60 min lacking enzymes. (B) Diagrammatic representation of a section of p/t-1 template strand DNA indicating the positions at which the three replicative proteins stall replication at the 10 min time point, 8-oxodG is indicated by O. (C) Graphical representation of replication stalling profiles induced by 8-oxodG for each of the replicative proteins, shown for each individual stall event and as total 8-oxodG induced arrest at the 10 min time point (raw data for each graph are provided in Figure 3—source data 1).*-Data for p41/p46 8-oxodG 32nt stall are not visible due to mirroring that of the dG control. (D) Effects of RPA and PCNA on both primer extension and TLS activity of PolB, PolD and the p41/p46 complex during a 15 min reaction with p/t-1 containing either dG or 8-oxodG.
-
Figure 3—source data 1
Qantification of gel bands arising from 8-oxodG induced stalling of the polymerisation enzymes from P. abyssi.
- https://doi.org/10.7554/eLife.45320.013

Primer extension under ‘standing start’ reaction conditions.
(A) Replicative enzyme activity when polymerisation is initiated from a pre-stalled start using p/t-3 containing either dG of 8-oxodG. The triangle above each gel image indicates time course of primer extension (time points taken at 0, 0.5, 1, 5, 10, 30 and 60 min). Primer+n compared to dG was calculated as formation of primer+n in 8-oxodG containing templates as a relative percentage of primer+n formation in dG containing templates. (B) Influence of RPA and PCNA on replicative enzyme activity when polymerisation is initiated from a pre-stalled start. Primer extension and translesion synthesis activity of PolB, PolD and the p41/p46 complex performed during a 15 min reaction.
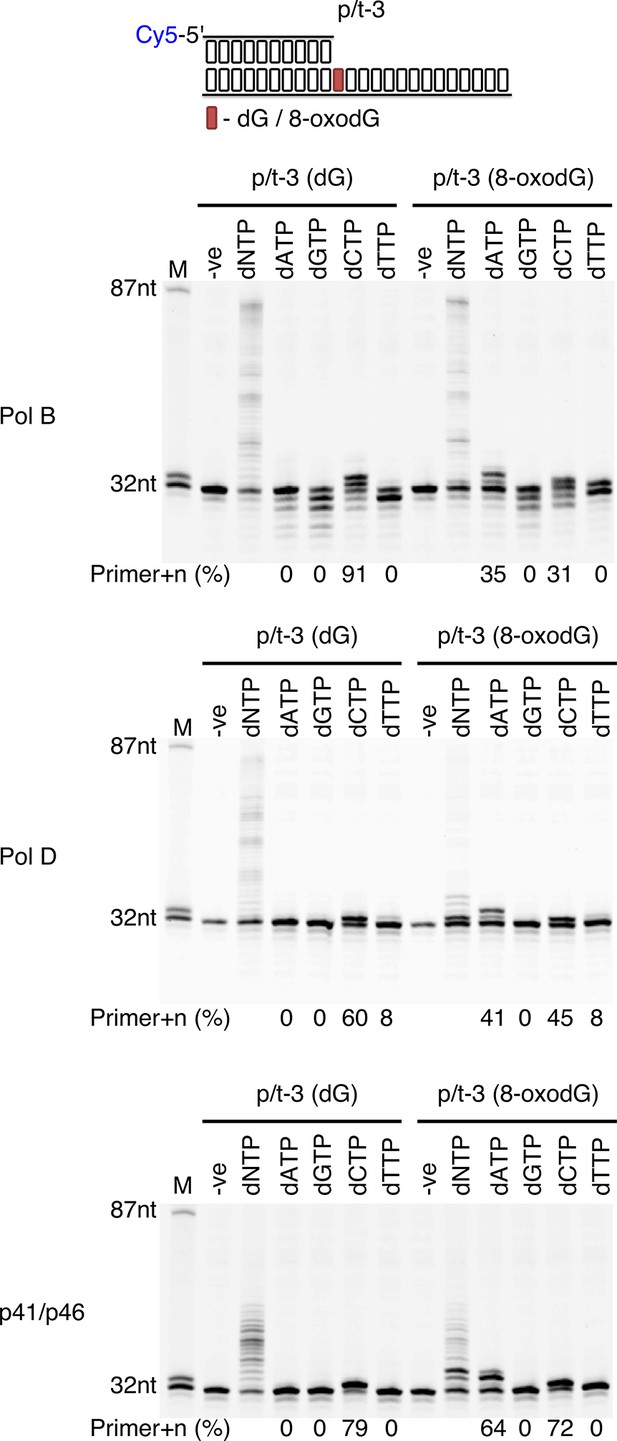
Single nucleotide incorporation opposite 8-oxodG by DNA polymerisation enzymes.
Single nucleotide incorporation reactions were performed for each of the three replicative enzymes to determine the accuracy of incorporation when incorporating a single nucleotide opposite either dG or 8-oxodG when located at the +1 position from the primer-template junctions (p/t-3). All reactions were left for 5 min, with the –ve control lacking replicative enzyme.
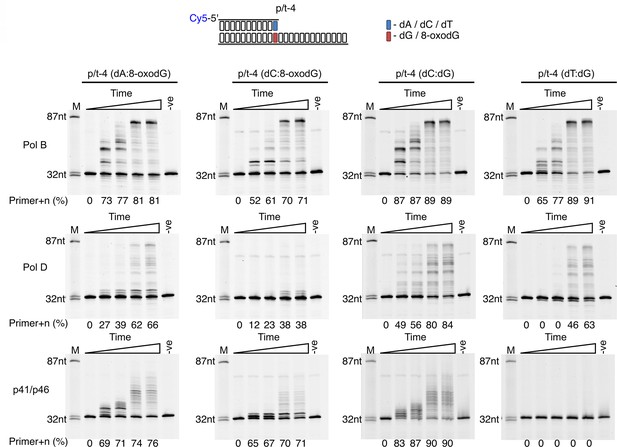
Extension of primers pre-base paired opposite 8-oxodG.
Primer-template extension reactions performed on p/t-4 containing either dG or 8-oxodG by PolB, PolD and the p41/p46 complex. The triangle above each gel image indicates time course of primer extension (time points taken at 0, 0.5, 1, 5 and 10 min). In all instances, the –ve samples were incubated for 60 min lacking enzymes.
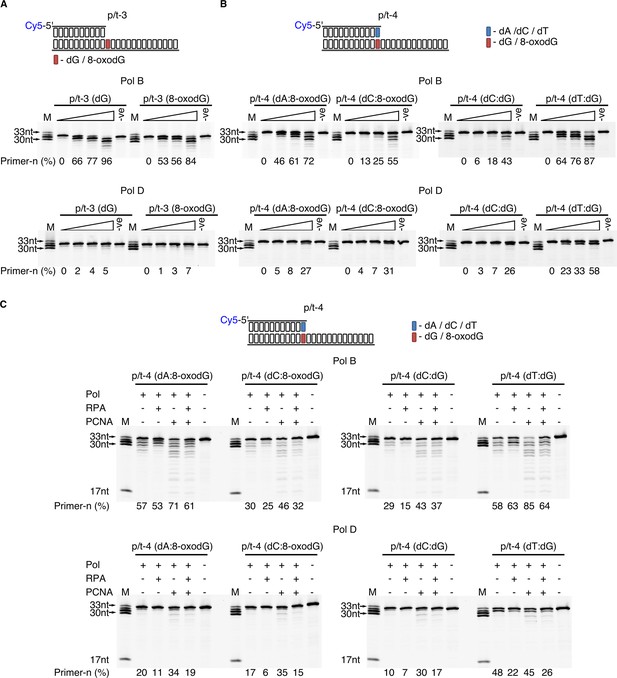
Effect of template strand 8-oxodG and 8-oxodG base paired termini on 3’−5’ exonuclease activity.
Exonuclease activity of PolB and PolD in the presence of dG and 8-oxoxdG when located in p/t-3,+1 nucleotides from the primer-template junction) (A) or when base paired with a complementary or non-complementary nucleotide in p/t-4, 0 nucleotide from the primer-template junction (B). In all instances, the triangle above the gel images denotes an exonuclease time-course with time points taken at 0, 0.5, 1 and 5 min. For all gels the –ve samples were incubated for 5 min lacking replicative enzyme. (C) Influence of RPA and PCNA on the exonuclease activity of PolB and PolD when encountering dG and 8-oxodG base paired with a complementary or non-complementary nucleotide (p/t-4) during a 5 min reaction.
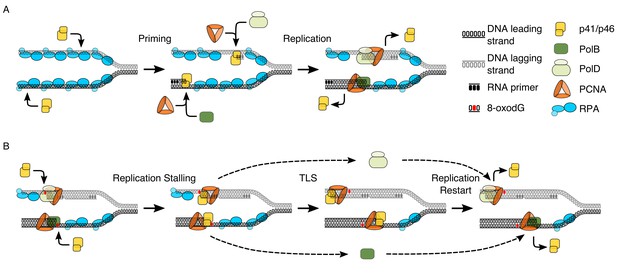
Model proposing the dual functionality of p41/p46 complex based on protein function and known interactions.
(A) Priming: p41/46 is recruited at the replication fork and engages in the synthesis of a short RNA primer in a manganese catalysed reaction on both the leading and lagging strand. On the leading strand switching to a catalytic magnesium ion occurs, resulting in incorporation of dNTPs and extension of the RNA primer by p41/p46 until the loading of PCNA and the recruitment of PolB displace it to initiate DNA synthesis. Synthesis of an RNA primer by p41/p46 leads to direct loading of PCNA and recruitment of PolD to initiate DNA synthesis on the lagging strand (Henneke et al., 2005; Le Breton et al., 2007). (B) TLS: DNA replication stalls upon the HiFi Pol active site encountering oxidative damage. Unable to bypass the damaged nucleobase, polymerase switching occurs, with p41/p46 recruited to the primer-template junction and retained by PCNA. RPA bound to the single stranded region of DNA stabilizes the phosphodiester backbone downstream of the damage, allowing p41/p46 to function as a TLS polymerase, replicating past the damaged nucleobase until it is eventually replaced with a processive HiFi DNA polymerase.
Tables
Rate of endogenous genomic 8-oxodG/106 dG in P. abyssi and E. coli genomes at different growth phases.
Steady-state level of 8-oxodG per 106 dG was calculated during the exponential and stationary growth phases. The number of 8-oxodG per 106 dG represents the average of triplicate experiments from two biological samples with the standard deviation (±) shown. ND means No Detectable (ND is assigned to values below the HPLC-EC-UV detection limit of 0.01 pmol of 8-oxodG). Raw data are presented in Table 1—Source data 1.
| Growth phase | 8-oxodG/106 dG | |
|---|---|---|
| E. coli | Exponential | ND |
| Stationary | ND | |
| P. abyssi | Exponential | 63.2 ± 4.6 |
| Stationary | 115.1 ± 5.8 |
-
Table 1—source data 1
Quantification of the steady-state level of 8-oxoguanosine in the genome of E.coli and P.abyssi.
- https://doi.org/10.7554/eLife.45320.004
Additional files
-
Supplementary file 1
Conformation of primer-templates (p/t) used in this study and their oligonucleotide sequences.
The location of the fluorescent labels is indicated by *.
- https://doi.org/10.7554/eLife.45320.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45320.017





