Cell non-autonomous functions of S100a4 drive fibrotic tendon healing
Figures
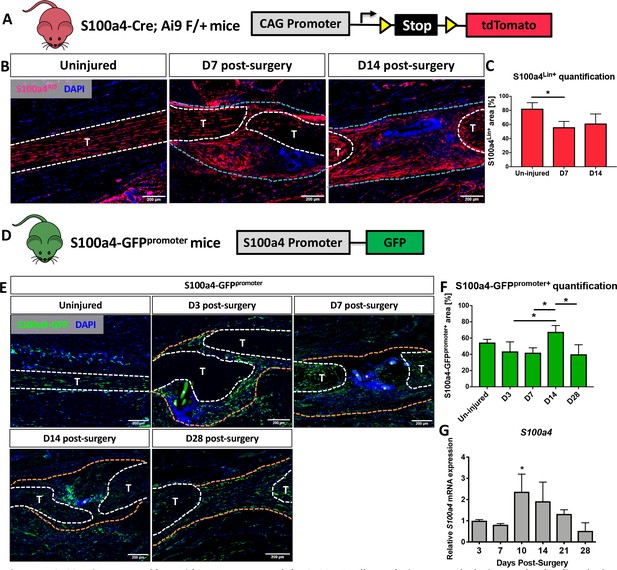
S100a4 is expressed by resident tenocytes and the S100a4+cell population expands during tendon healing.
(A and B) S100a4-Cre; Rosa-Ai9 reporter mice demonstrate efficient targeting of resident tendon cells. Following injury, the S100a4-lineage (S100a4Lin+) population expands, with S100a4Lin+ cells in the native tendon stubs and the bridging scar tissue at D7 and D14 post-surgery. Tendons are outlined in white, and bridging granulation tissue outlined in blue. (C) Quantification of S100a4Lin+ area over time. (*) indicates p<0.05 (1-way ANOVA). (D) The S100a4-GFPpromoter construct identifies cells actively expressing S100a4 (S100a4-GFPpromoter+). (E) A subpopulation of resident tenocytes is S100a4-GFPpromoter+ at baseline, and the S100a4-GFPpromoter+ population increases following injury, with S100a4-GFPpromoter+ cells observed in the bridging scar tissue and native tendon ends through D28 post-surgery. Tendons are outlined in white, and bridging granulation tissue outlined in orange, (*) identifies sutures. (F) Quantification of the S100a4-GFPpromoter+ area over time. (*) indicates p<0.05 (1-way ANOVA). (G) qPCR analysis of S100a4 during tendon healing demonstrates peak S100a4 expression at D10, followed by a progressive decline through D28 (n = 3 per time-point). (*) indicates p<0.05 vs. D3 repair (1-way ANOVA). Data were normalized to expression in D3 repairs, and the internal control β-actin.

S100a4+cells are found in the healthy and healing Achilles tendon.
S100a4-GFPPromoter+ cells are observed in the native Achilles tendon, and a S100a4+ population persists following complete transection and repair of the Achilles tendon at D14 post-surgery.
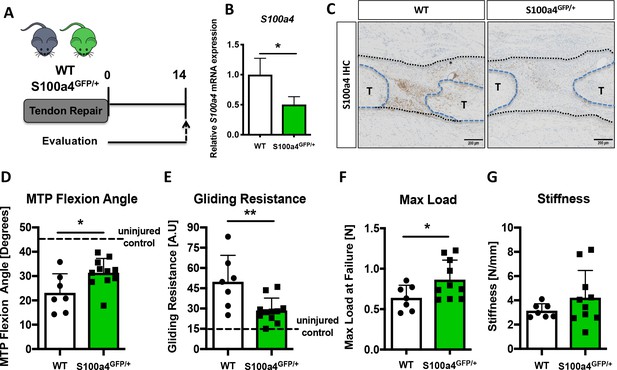
S100a4 haploinsufficiency promotes regenerative, mechanically superior tendon healing.
(A) S100a4GFP/+ haploinsufficient and wild type (WT) littermates underwent transection and repair of the FDL tendon, and tendons were harvested at D14 post-surgery. (B) S100a4 mRNA expression was reduced by 50% in S100a4GFP/+ tendon repairs, relative to WT (n = 3 per group). (C) A substantial reduction in S100a4 protein expression was observed in S100a4GFP/+ tendon repairs, relative to WT. Tendon ends are outlined in blue and bridging scar tissue outlined in black (n = 3–4 per group). (D–G) At D14, MTP Flexion Angle was significantly increased in S100a4GFP/+ repairs (D), and Gliding Resistance was significantly decreased in S100a4GFP/+ repairs (E). Max load at failure was significantly improved in S100a4GFP/+ repairs (F), while no change in Stiffness was observed between genotypes (G) (n = 7–10 per group). (*) indicates p<0.05, (**) indicates p<0.01 between genotypes, n = 7–10 for (D–G) (un-paired t-test).

S100a4 haploinsufficiency does not alter gliding function or mechanical properties of un-injured tendons.
No changes in (A) MTP Flexion Angle, (B) Gliding Resistance, (C) Max load at failure, or (D) Stiffness were observed between WT and S100a4GFP/+ un-injured contralateral control FDL tendons. n = 11–13, (un-paired t-test).
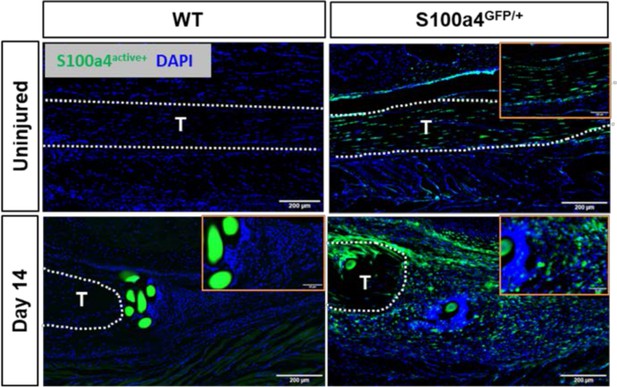
S100a4GFP/+mice permit tracing of S100a4 haploinsufficient cells.
To determine if S100a4 haploinsufficiency altered the S100a4+ population during healing, fluorescent imaging of uninjured and D14 repairs from S100a4GFP/+ and S100a4+/+ (WT) mice were analyzed. No GFP expression was observed in S100a4+/+ WT mice either at baseline or at D14 post-surgery. In contrast, knock-down of S100a4 does not alter the S100a4GFP+ resident tendon cell population, or the expansion of the S100a4GFP+ population at D14 post-surgery. Orange insets identify high-power magnification images.
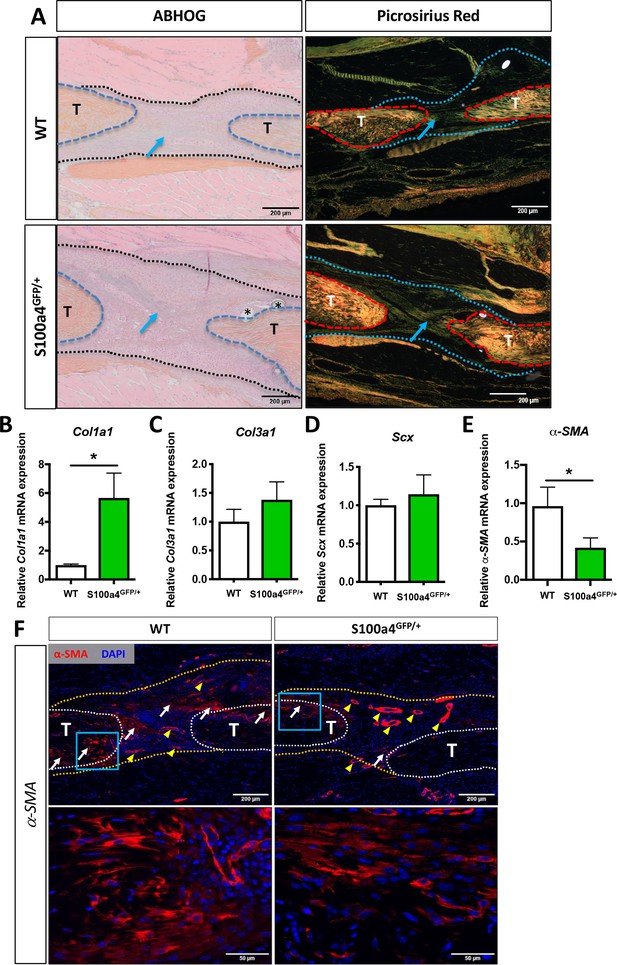
S100a4 haploinsufficiency enhances deposition of a mature Collagen matrix and reduced myofibroblast content.
(A) ABH/OG and picrosirius red staining demonstrate an increase in mature collagen fibers (blue arrows) bridging the tendon ends in S100a4GFP/+ repairs compared to WT littermates (n = 3–4 per group) (*) indicate sutures. (B–D) S100A4GFP/+ tendons expressed significantly more Col1a1 mRNA (B), while transcript levels of Col3a1 (C) and Scx (D) were unaffected by S100a4 haploinsufficiency. (*) indicates p<0.05 (un-paired t-test), n = 3 per group. (E and F) α-SMA mRNA expression was significantly decreased in S100a4GFP/+ repairs (E) (n = 3 per group), while a substantial reduction in α-SMA protein expression was observed in S100a4GFP/+, relative to WT, using immunofluorescence (F). White arrows indicate areas of α-SMA+ cells in the healing tissue, yellow arrowheads denote α-SMA staining of vessels, blue boxes indicate location of higher magnification images.
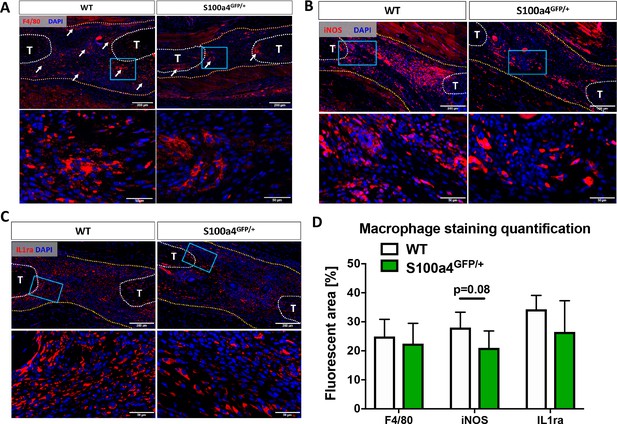
S100a4 haploinsufficiency alters the macrophage response to tendon injury.
(A) F4/80 staining demonstrates decreased macrophage content in the healing tendon of S100a4GFP/+ repairs at D14. White arrows identify concentrated areas of macrophages. (B) Expression of the M1 macrophage marker iNOS is markedly reduced in S100a4GFP/+ repairs at D14. (C) Expression of the M2 macrophage marker IL1ra is not different between WT and S100a4GFP/+ repairs at D14. Tendon ends are outlined in white, scar tissue is outlined in yellow, blue boxes indicate location of higher magnification images (n = 4 per group). (D) The percent area of F4/80+, iNOS+ and IL1ra+ staining, normalized to tissue area was quantified (n = 4) (un-paired t-test).
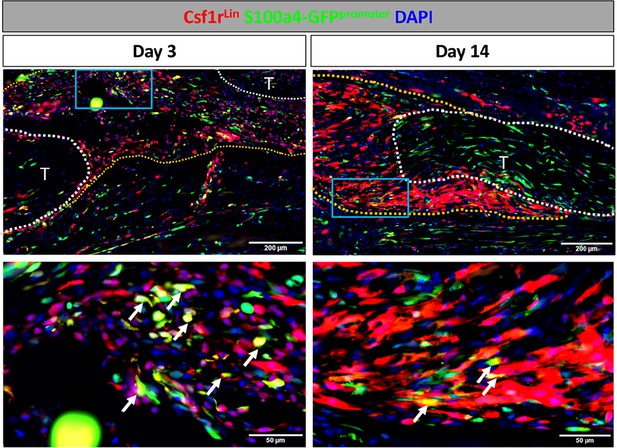
S100a4 is expressed by macrophages during early tendon healing.
To identify S100a4+ macrophages, Csf1r-iCre; Rosa-Ai9; S100a4-GFPpromoter+ mice were induced with tamoxifen. Tmx was given on D0-2 for mice harvested on D3, and D0-2 and every 48 hr thereafter until harvest for samples harvested at D14. On D3 several macrophages actively express S100a4 (white arrows). By D14 the presence of Csfr1Lin+ cells increased, but very few cells actively expressed S100a4 (white arrows).
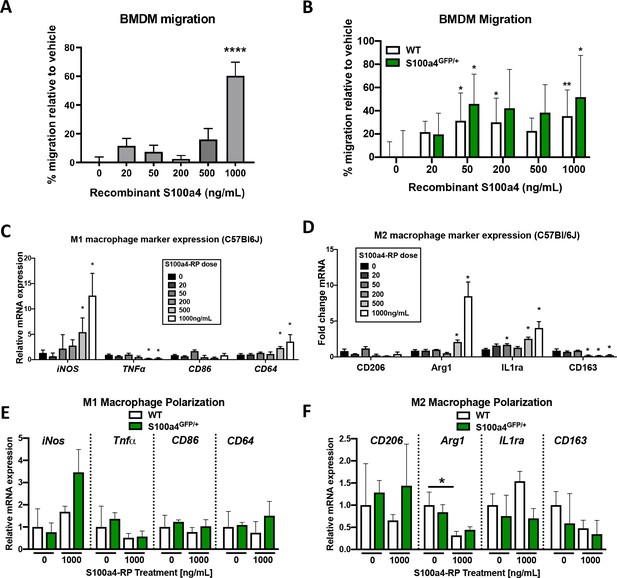
S100a4 promotes macrophage migration and alters polarization.
(A) S100a4 promotes migration of C57BL/6J bone marrow derived macrophages (BMDMs). (****) Indicates p<0.0001 vs. vehicle treated cells (1-way ANOVA). (B) No change in migration was observed in vehicle treated WT and S100a4GFP/+ BMDMs, while significant increases in migration were observed in WT and S100a4GFP/+ BMDM treated with 50 ng/mL and 1000 ng/mL S100a4-RP. (*) indicates p<0.05, (**) indicates p<0.01 vs. genotype-matched vehicle treated cells (2-way ANOVA). (C and D) Following treatment with S100a4-RP (20–1000 ng/mL), significant increases in M1 polarization markers (iNos, CD64) were observed relative to vehicle treated C57BL/6J BMDMs, as was a significant decrease in TNFα, and no change in CD86 expression (C). Significant increases in M2 markers Arg2 and IL1ra were seen with S100a4-RP treatment, while a decrease in CD163 expression, and no change in CD206 was also observed in C57BL/6J BMDMs (D). (*) indicates p<0.05 between vehicle and S100a4-RP treatment (1-way ANOVA). (E and F) Expression of M1 (E) and M2 (F) macrophage markers were not significantly different between WT and S100a4GFP/+ BMDMs in vehicle treated or upon treatment with 1000 ng/mL S100a4-RP. (*) indicates p<0.05 vs. genotype-matched vehicle treated cells (2-way ANOVA). (n = 3 per treatment).

Tendon cell S100a4 haploinsufficiency does not alter tenogenic and matrix gene expression or proliferation but enhances migration.
(A) qPCR analysis primary tendon cells from WT and S100a4GFP/+ mice. Expression of S100a4 is significantly reduced (~50%) in S100a4GFP/+ tendon cells, relative to WT. No changes in tenogenic genes Scx, Tnmd and Mkx. (*) indicates p<0.05 vs. expression in WT cells. (B) No changes in expression of matrix genes Col1a1, Col3a1 or Fn are observed between WT and S100a4GFP/+ tendon cells. Data are normalized to WT expression and β-actin. (un-paired t-test). (C) No changes in proliferation were observed between WT and S100a4GFP/+ tendon cells (2-way ANOVA). (D). Cell migration was assessed by measuring closure of a scratch wound. Data are plotted as % of initial scratch area. Closure was significantly increased in S100a4GFP/+ tendon cells at 24 hr (2-way ANOVA).
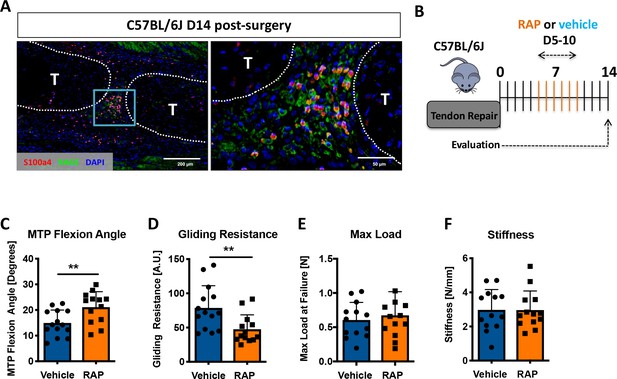
Inhibition of S100a4 signaling via RAGE antagonism improves tendon healing.
(A) Co-immunofluorescence demonstrated co-localization of S100a4 and its putative receptor RAGE in the healing tendon (n = 3). (B) C57Bl/6J mice were treated with either RAP or vehicle, via i.p. injection from D5-10 post-surgery, and harvested at D14 for functional testing. (C–F) At D14 RAP treatment significantly improved measures of gliding function relative to vehicle, with a (C) significant increase in MTP Flexion Angle, and (D) a significant decrease in Gliding Resistance. No change in (E) Max load at failure, or (F) Stiffness was observed between treatments (n = 13 per group). (**) indicates p<0.01 between treatments (un-paired t-test).
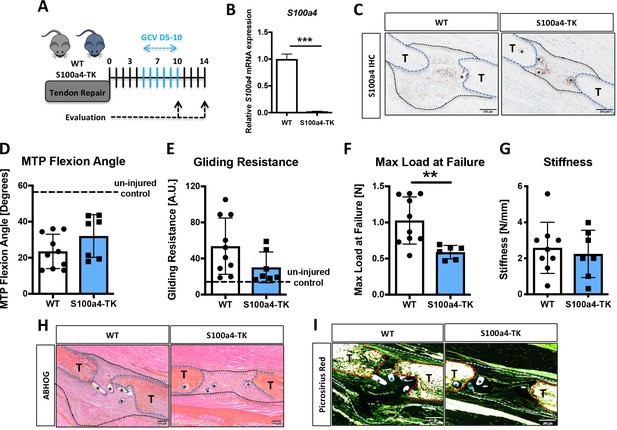
Delayed depletion of S100a4+cells impairs restoration of mechanical properties and alters matrix deposition.
(A) WT and S100a4-TK mice were treated twice daily with ganciclovir (GCV) from D5-10 post-surgery. (B) S100a4+ cell depletion results in a 91% reduction in S100a4 mRNA at D10 post-surgery (n = 3). (C) A substantial reduction in S100a4 protein expression was observed S100a4-TK repairs, relative to WT. Tendon is outlined in blue, scar tissue is outlined in black and (*) identify sutures (n = 4). (D–G) At D14 no change in MTP Flexion Angle (D) and Gliding Resistance (E) were observed between WT and S100a4-TK repairs. (F) Max load at failure was significantly reduced following S100a4-cell depletion, while no change in Stiffness was observed (G) (n = 7–10), (**) indicates p<0.01 (un-paired t-test). (H and I) Morphologically, (H) ABH/OG and (I) Picrosirius staining demonstrate reduced matrix deposition bridging the tendon ends in the S100a4-TK repairs, relative to WT. (*) Indicates sutures.
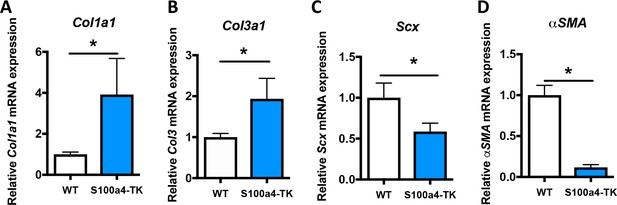
S100a4+cell depletion alters expression of matrix, tenogenic and myofibroblast-associated genes.
qPCR analyses demonstrated significant increases in (A) Col1a1, and (B) Col3a1 expression, while (C) Scx and (D) α-SMA expression levels were significantly reduced in S100a4-TK, relative to WT. Data were normalized to expression in WT samples and the internal control β-actin. (*) Indicates p<0.05, (***) indicates p<0.001, (n = 3 per group) (un-paired t-test).
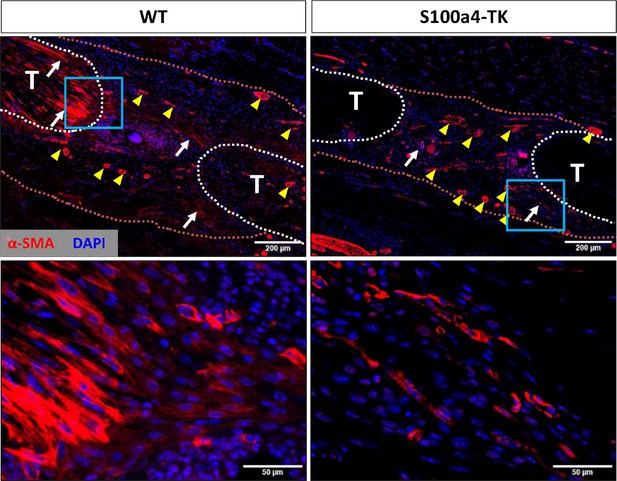
S100a4+cell depletion reduces α-SMA+myofibroblast content during healing.
At D14 post-surgery, abundant α-SMA+ myofibroblasts (red) were observed in WT repairs. In contrast, α-SMA+ myofibroblast content was markedly reduced in S100a4-TK (D5-10) repairs. White arrows indicate areas of α-SMA+ cells in the healing tissue, while yellow arrowheads denote α-SMA staining of vessels. Tendons are outlined in white and scar tissue outlined in red.
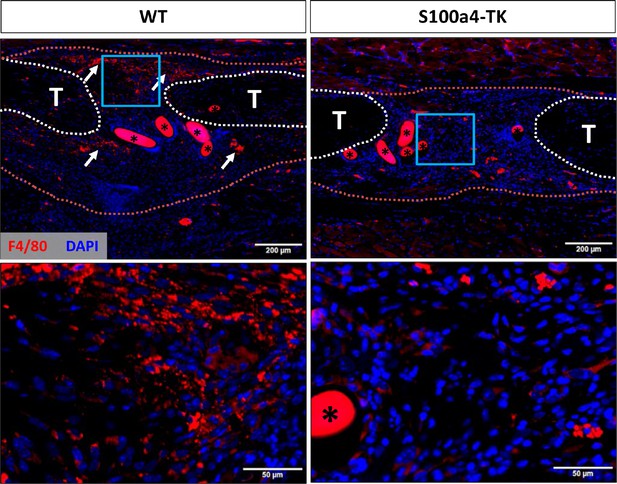
S100a4+cell depletion reduces macrophage content during healing.
At D14 post-surgery abundant F4/80+ macrophages (red) were observed in WT repairs. In contrast, F4/80+ macrophage content was markedly reduced in S100a4-TK (D5-10) repairs. White arrows identify concentrated areas of macrophages. Tendon ends are outlined in white, while scar tissue is outlined in red. (*) indicates sutures.
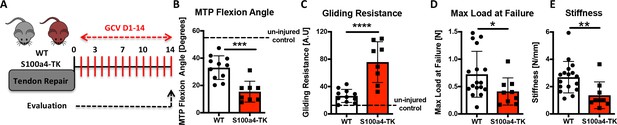
Sustained ablation of S100a4+impairs restoration of gliding function and mechanical properties.
(A) WT and S100a4-TK mice were treated with GCV from D1-14 post-surgery to ablate proliferating S100a4+ cells. At D14 (B) MTP Flexion Angle was significantly reduced, and (C) Gliding Resistance was significantly increased in S100a4-TK repairs, relative to WT. (D) A non-significant decrease in Max load at failure and (E) a significant reduction in Stiffness were observed in S100a4-TK repairs (n = 8–11). (*) indicates p<0.05, (**) indicates p<0.01 (un-paired t-test).
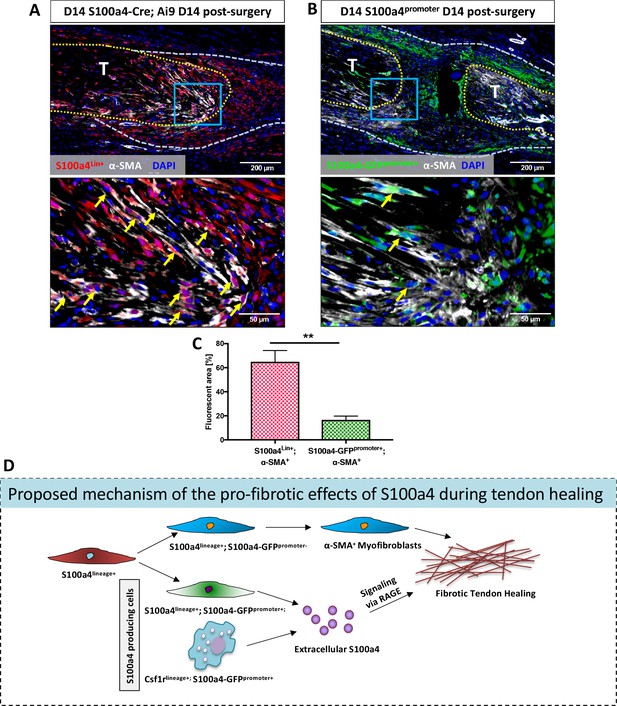
S100a4-lineage cells lose S100a4 expression during the transition to α-SMA+myofibroblasts.
(A) Co-localization of Red Fluorescent Protein (S100a4Lin+ cells; red) and the myofibroblast marker α-SMA (white) demonstrated abundant co-localization (yellow arrows) during tendon healing. (B) Minimal co-localization of α-SMA (white) and cells actively expressing S100a4 (S100a4-GFPpromoter+; green) was observed during healing (n = 3). (C) Quantification of the percent α-SMA+ area that is also S100a4Lin+ (red and white bar) or S100a4-GFPpromoter+ (green and white bar) at D14 (n = 3–4 per group) (**) indicates p<0.01 between groups (un-paired t-test). (D) Schematic representation of the proposed cell non-autonomous signaling functions of S100a4 in fibrotic healing, as well as cell fate of S100a4-lineage cells. The identities and discrete functions of specific populations of S100a4+ cells remains to be determined.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus. musculus) | B6.Cg-Tg(S100a4-EGFP)M1Egn/YunkJ (S100A4-GFPpromoter) | Jackson Laboratory | Stock #: 012893 RRID: MGI:4819362 | |
| Genetic reagent (M. musculus) | B6.Cg-Tg(S100a4-TK)M31Egn/YunkJ (S100a4-TK) | Jackson Laboratory | Stock #: 012902 RRID:MGI:4454768 | |
| Genetic reagent (M. musculus) | B6.129S6-S100a4tm1Egn/YunkJ (S100a4GFP/+) | Jackson Laboratory | Stock #: 012904 RRID:MGI:4819358 | |
| Genetic reagent (M. musculus) | BALB/c-Tg(S100a4-cre)1Egn/YunkJ (S100a4-Cre) | Jackson Laboratory | Stock #: 012641 RRID:MGI:4454332 | |
| Genetic reagent (M. musculus) | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (ROSA-Ai9) | Jackson Laboratory | Stock #: 007909 RRID:MGI:3809523 | |
| Genetic reagent (M. musculus) | Tg(Csf1r-Mer-iCre-Mer)1Jwp (Csf1r-iCre) | Jackson Laboratory | Stock #: 019098 RRID:IMSR_JAX:019098 | |
| Genetic reagent (M. musculus) | C57BL/6J | Jackson Laboratory | Stock #: 000664 RRID:MGI:3028467 | |
| Antibody | anti-RAGE (mouse monoclonal) | Santa Cruz Biotechnology | Cat. #: sc-365154 RRID:AB_10707685 | 1:100 |
| Antibody | anti-F4/80 (goat polyclonal) | Santa Cruz Biotechnology | Cat. #: sc-26642 RRID:AB_2098333 | 1:500 |
| Antibody | anti-GFP (goat polyclonal) | Abcam | Cat. #: ab6673 RRID:AB_305643 | 1:5000 |
| Antibody | anti-RFP (rabbit polyclonal) | Abcam | Cat. #: ab62341 RRID:AB_945213 | 1:500 |
| Antibody | anti-S100a4 (rabbit monoclonal) | Abcam | Cat. #: ab197896 RRID:AB_2728774 | 1:20000 |
| Antibody | anti-iNOS (rabbit polyclonal) | Abcam | Cat. #: ab15323 RRID: AB_301857 | 1:100 |
| Antibody | Anti-ILIRa (rabbit monoclonal) | Abcam | Cat. #: ab124962 RRID:AB_11130394 | 1:10000 |
| Antibody | anti-alpha-SMA-Cy3 (mouse monoclonal) | Sigma-Aldrich | Cat. #: C6198 RRID: AB_476856 | 1:250 |
| Antibody | Donkey anti-rabbit AlexaFluor594 secondary | Jackson ImmunoResearch | Cat. #: 711-585-152 RRID: AB_2340621 | 1:200 |
| Antibody | Donkey anti-rabbit 647 secondary | Jackson ImmunoResearch | Cat. #: 711-605-152 RRID: AB_2492288 | 1:200 |
| Antibody | Donkey anti-rabbit Rhodamine-Red-X | Jackson ImmunoResearch | Cat. #: 711-296-152 RRID:AB_2340614 | 1:100 |
| Antibody | Donkey anti-goat 488 secondary | Jackson ImmunoResearch | Cat. #: 705-546-147 RRID: AB_2340430 | 1:200 |
| Antibody | Goat anti-mouse AlexaFluor488 secondary | ThermoFisher | Cat. #: A11029 RRID:AB_138404 | 1:1000 |
| Chemical compound, drug | Nucleoside analog ganciclovir (GCV) | TSZCHEM | Cat #: 82410-32-0 | 75 mg/kg |
| Peptide, recombinant protein | RAGE Antagonist Peptide (RAP) | MilliporeSigma | Cat. #: 553031 | 100 ug (peptide) |
| Peptide, recombinant protein | Human S100a4 Recombinant protein | LSBio | Cat #: G1305 | 20–1000 ng/mL (recombinant protein) |
| Commercial kit | Rabbit polymer kit | Vector Laboratories | Cat #: MP-7401 | |
| Software | OlyVIA software | Olympus (https://www.olympus-lifescience.com/en/support/downloads/) | RRID:SCR_016167 | Version 2.9 |
| Software | ImageJ software | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software | GraphPad Prism software | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 8.0.0 |
qPCR Primer Sequences
https://doi.org/10.7554/eLife.45342.019| Gene | Sequence (5'- > 3') | Reference | |
|---|---|---|---|
| Actb | Fwd | AGATGTGCATCAGCAAGCAG | NM_007393.5 |
| Rev | GCGCAAGTTAGGTTTTGTCA | ||
| S100a4 | Fwd | AAGCTGAACAAGACAGAGCTCAAG | NM_011311.2 |
| Rev | GTCCTTTTCCCCAGGAAGCTA | ||
| Fn | Fwd | CGAGGTGACAGAGACCACAA | NM_001276413.1 |
| Rev | CTGGAGTCAAGCCAGACACA | ||
| Tnmd | Fwd | TGTACTGGATCAATCCCACTCT | NM_022322.2 |
| Rev | GCTCATTCTGGTCAATCCCCT | ||
| Scx | Fwd | TGGCCTCCAGCTACATTTCT | NM_198885.3 |
| Rev | TGTCACGGTCTTTGCTGAAC | ||
| Mkx | Fwd | CACCGTGACAACCCGTACC | NM_177595.4 |
| Rev | GCACTAGCGTCATCTGCGAG | ||
| Col1a1 | Fwd | GCTCCTCTTAGGGGCCACT | NM_007742.4 |
| Rev | CCACGTCTCACCATTGGGG | ||
| Col3a1 | Fwd | ACGTAGATGAATTGGGATGCAG | NM_009930.2 |
| Rev | GGGTTGGGGCAGTCTAGTG | ||
| Acta2 | Fwd | GAGGCACCACTGAACCCTAA | NM_007392.3 |
| Rev | CATCTCCAGAGTCCAGCACA | ||
| iNOS | Fwd | CAGAGGACCCAGAGACAAGC | NM_001313921.1 |
| Rev | TGCTGAAACATTTCCTGTGC | ||
| TNFa | Fwd | AACTGTAAGCGGGGCAATCA | NM_013693.3 |
| Rev | CCCCTTTCCTCCCAAACCAA | ||
| Cd86 | Fwd | TCTCCACGGAAACAGCATCT | NM_019388.3 |
| Rev | CTTACGGAAGCACCCATGAT | ||
| Cd64 | Fwd | TCCTTCTGGAAAATACTGACC | NM_010186.5 |
| Rev | GTTTGCTGTGGTTTGAGACC | ||
| Cd206 | Fwd | CAGGTGTGGGCTCAGGTAGT | NM_008625.2 |
| Rev | TGTGGTGAGCTGAAAGGTGA | ||
| Arg1 | Fwd | AGGAACTGGCTGAAGTGGTTA | NM_007482.3 |
| Rev | GATGAGAAAGGAAAGTGGCTGT | ||
| IL1ra | Fwd | GCATCTTGCAGGGTCTTTTC | NM_001159562.1 |
| Rev | GTGAGACGTTGGAAGGCAGT | ||
| Cd163 | Fwd | TCCACACGTCCAGAACAGTC | NM_001170395.1 |
| Rev | CCTTGGAAACAGAGACAGGC |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45342.020





