Translation affects mRNA stability in a codon-dependent manner in human cells
Figures
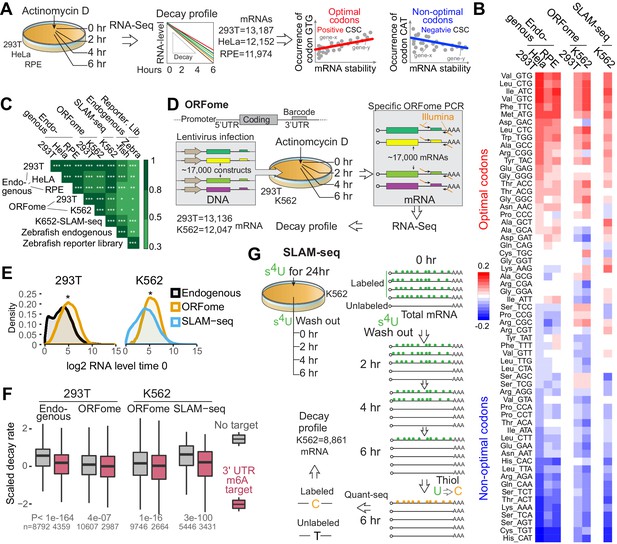
Human mRNA decay correlate with codon composition in human lines.
(A) Scheme of the mRNA endogenous decay profiles approach in 293T, HeLa and RPE cells. RNA-seq was performed at different time after blocking transcription by Actinomycin D treatment. The codon stabilization coefficient (CSC) is calculated as the Pearson correlation coefficient between the occurrence of each codon and mRNA stability. (B) Heatmap showing the codon stability coefficient (CSC) calculated in the indicated cell lines and approaches used to measure mRNA stability. (C) Heatmap showing the Spearman rank correlations between CSC calculated in different human cells lines and using different methods to calculate mRNA stability. CSC calculated in zebrafish embryos with two independent methods were included (Bazzini et al., 2016). *p<0.05, **p<0.01 and ***p<1e-16. (D) Diagram illustrating the ORFome decay profiles approach. Cells were infected with lentivirus carrying approximately 17,000 expression vectors containing different coding sequences but sharing the same promoter, 5’ and 3’UTRs. Each vector contains a 20-nucleotide long unique barcode in the 3’UTR. Infected cells were selected by puromycin treatments. RNA was isolated at different time after blocking transcription by Actinomycin D. Specific RNA-seq libraries were generated amplifying the shared sequences surrounding the barcodes and mRNA ORFome decay was calculated based on the sequences. (E) Density distribution of the mRNA level in 293T or K562 cells using the indicated techniques. Two-sample Kolmogorov-Smirnov test is used, *p<1e-50. (F) Boxplot showing the scaled mRNA decay of previously defined m6A targets mRNAs and as control, mRNAs that were not defined at m6A targets in the indicated cell line and using the indicated method to calculate decay. A linear model is used to estimate the difference. Number of genes (n) and p value KS test indicated. (G) Schematic representation of the SLAM-seq. Cells were fed with s4U every 3 hr for 24hs hours. Orthogonal-chemistry-based RNA sequencing was preformed after washing out the s4U. Decay profiles were calculated based on the chase of the label/unlabeled mRNAs (T to C conversion).
-
Figure 1—source data 1
mRNA stability profiles used in Figure 1.
- https://doi.org/10.7554/eLife.45396.005
-
Figure 1—source data 2
CSC scores from Figure 1B.
- https://doi.org/10.7554/eLife.45396.006
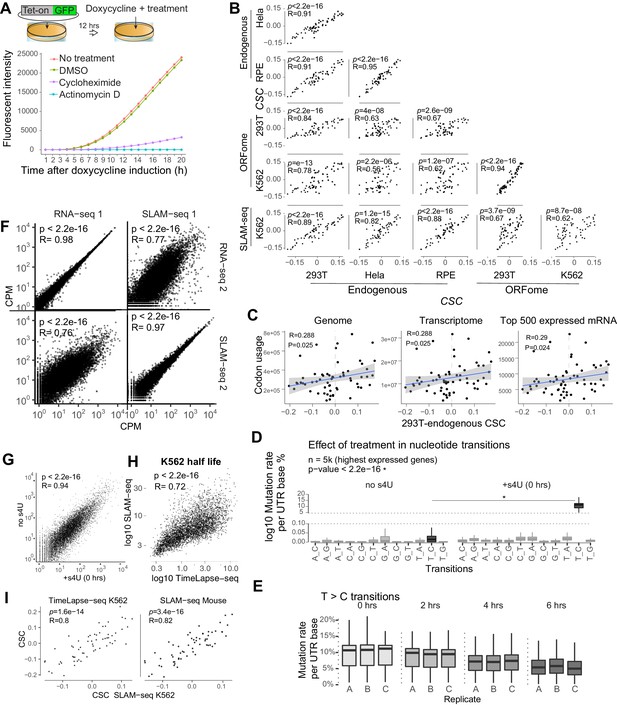
Human and mouse mRNA decay correlate with codon composition in human lines.
(A) Drug condition analysis for actinomycin D and Cycloheximide in 293T cells. Transfected cells with tet-on GFP, were treated with different drugs and induced with Doxycycline at time 0 hr. Then cells were under time-course fluorescent detection. Cycloheximide (2 μg/ml) repressed translation, with low GFP level, Actinomycin D(5 μg/ml) blocked transcription, with no GFP expression detected. (B) Scatter plot for CSC scores from different human cell lines and/or methods measuring mRNA decay. (C) Scatter plots showing 293T endogenous CSC scores comparing to codon usage calculated from the genome, transcriptome (each gene with the weight of RPKM) or the top 500 highly expressed genes. Spearman correlation is used, p value and R value is indicated. (D) Boxplot showing the ratio of nucleotide transition without and with s4U treatment using the top five thousand expressed mRNA, only T_C transition gets significant increase after labeling, indicating the SLAM-seq labeling works. Wilcox rank sum test is used, p.value indicated. (E) Boxplot showing the T to C transition in each triplicate over time. The ratio decreased with time for the chasing period. (F) Scatter plot for mRNA-seq with regular RNA-seq and SLAM-seq in duplicates. Pearson correlation and P-values indicated. (G) Scatter plot for mRNA level with and without s4U treatment, s4U treatment did not influence gene expression. Pearson correlation and P-value indicated. (H) Scatter plot showing mRNA half-life of endogenous genes in K562 cell measured with two different methods, TimeLapse-seq (Schofield et al., 2018), and our SLAM-seq. Pearson correlation and p-value indicated. (I) Scatter plot showing CSC scores from our SLAM-seq data in K562 cells, published dataset for TimeLapse-seq in K562 (Schofield et al., 2018), and published SLAM-seq data in mouse embryonic stem cell (Herzog et al., 2017). Pearson correlation and p-value indicated.
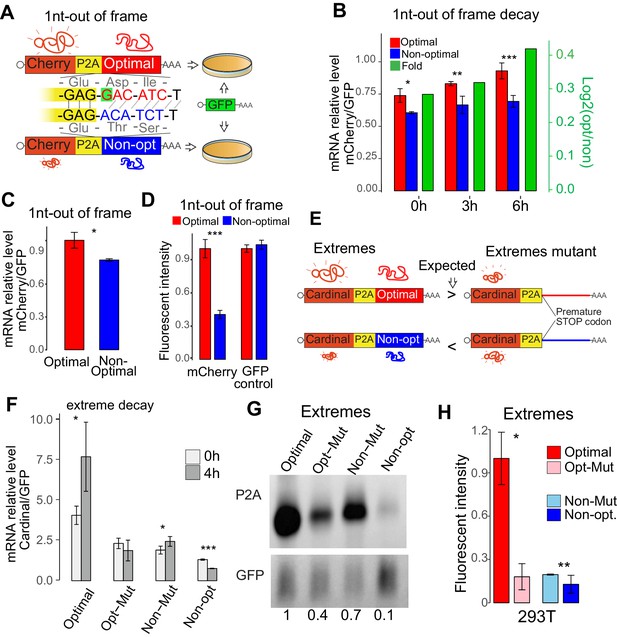
Codon identity affects mRNA stability in human lines.
(A) Scheme of the 1nt-out of frame reporters: two mRNAs that differ in the codon composition due to a single nucleotide (G in red, highlight with green) which creates a frameshift. The encoding mCherry fluorescent protein was followed by a cis-acting hydrolase elements (P2A) and then by the coding region enriched in optimal or non-optimal codons due to the frame shift. P2A causes ribosome skipping, therefore, the mCherry is not fused to the optimal or non-optimal encoded proteins. The reporter pairs were co-transfected with a vector encoding for GFP as internal control. (B) The 1nt-out of frame reporter enriched in optimal codons displayed higher stability than its non-optimal counterpart. Cells are treated with Actinomycin D 24hs after transfection, samples are collected at different timepoint to measure RNA level after blocking transcription, qPCR result is normalized with GFP internal control. *p<0.05, **p<0.01 and ***p<0.005 Unpaired t.test for all the panels. (C) qRT-PCR results showing that the 1nt-out of frame reporter enriched in optimal codons present higher level of RNA than its non-optimal counterpart 24 hr post-transfection. (D) The 1nt-out of frame reporter enriched in optimal codons displayed higher mCherry intensity than its non-optimal counterpart measure by cytometry. None significant differences were observed in the internal GFP control. (E) Scheme of the ‘Extreme’ reporters. Pairs of mRNA differing only in single mutation causing a premature stop codon. Each mRNAs contain Cardinal fluorescent protein coding region followed by a cis-acting hydrolase elements (P2A) and coding region enriched in optimal or non-optimal codons. Their counterparts (referred as mutants) contain a single mutation that cause a premature stop codon upstream the region enriched in optimal or non-optimal codons. Therefore, the translational competent reporter enriched in optimal codons should evidence higher level of expression that its mutant counterpart. And the reporter in non-optimal mRNA should express less than its counterpart translational incompetent. (F) qRT-PCR results showing relative stability of extreme reporters. Cells are treated with Actinomycin D 24hs after transfection, samples are collected at different timepoint to measure RNA level after blocking transcription, GFP is used as internal control to normalize qPCR result. The extreme translation-competent reporter enriched in optimal codons presented higher stability than its translation deficient counterpart (Mut). And the translation-competent reporter enriched in non-optimal codon was more unstable than its translation deficient counterpart (Mut). (G) Northern-blot showing that the extreme reporter enriched in optimal codons presented higher RNA level that its mutant counterpart. Contrary, the mRNA enriched in non-optimal codons present lower mRNA level than is mutant counterpart. GFP mRNA signal was used as internal control. Relative normalized quantifications are indicated. (H) The extreme reporter enriched in optimal codons displayed higher fluorescent intensity than its mutant counterpart measure by cytometry. The reporter enriched in non-optimal codons evidenced lower fluorescent intensity than its mutated pair.
-
Figure 2—source data 1
Reporter sequences and oligos used for Figure 2.
- https://doi.org/10.7554/eLife.45396.009
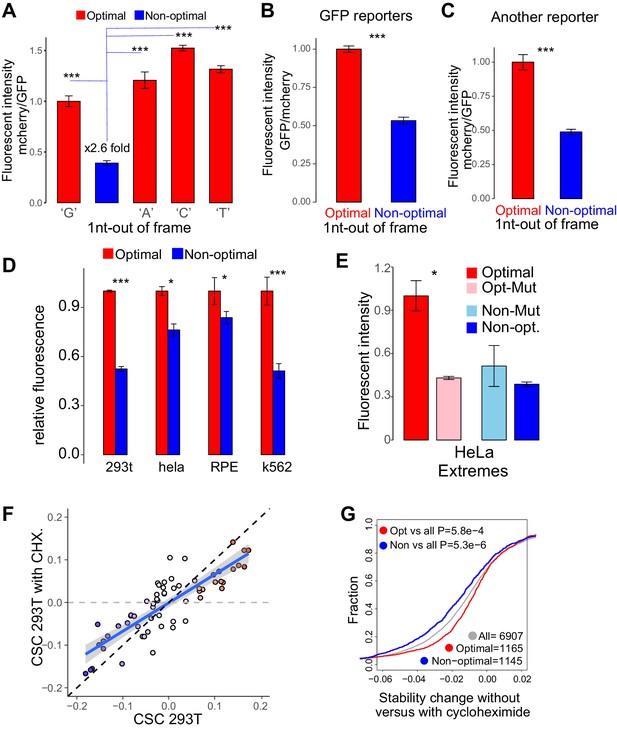
Codon identity affects mRNA stability in human lines in a translation-dependent manner.
(A) The 1nt-out of frame reporters enriched in optimal codons present higher mCherry intensity than its non-optimal counterpart independent of the nucleotide (G,A,C,T) used to introduce the frameshift. *p<0.05, **p<0.01 and ***p<0.005 Unpaired t.test for all the panels. (B) The 1nt-out of frame reporter with GFP (replacing mcherry) shows optimality effects at fluorescent intensity. (C) Another pair of 1nt-out of frame reporter, with totally different sequence, shows optimality effects at fluorescent intensity. (D) The 1nt-out of frame reporter shows optimality when we transfected into 293T, HeLa, RPE and K562 cells, with optimal reporter shows higher fluorescent intensity than non-optimal reporter (E) The extreme reporter enriched in optimal codons displayed higher fluorescent intensity than its mutant counterpart. And the reporter enriched in non-optimal codons evidenced lower fluorescent intensity than its mutated pair, when transfected into HeLa cell. (F) Scatter plot for CSC in 293 T cells, with and without Cycloheximide. Every circle is a codon and the color indicates CSC in 293T, linear regression with confident level is indicated with blue line, black dashed line is: y = x. (G) Cumulative distribution of mRNA stability change with and without Cycloheximide from mRNAs enriched in either optimal or non-optimal codons. mRNAs enriched in optimal became less stable in the presence of Cycloheximide and mRNAs enriched in non-optimal codons more stable. P-values indicated, Wilcoxon rank-sum test.
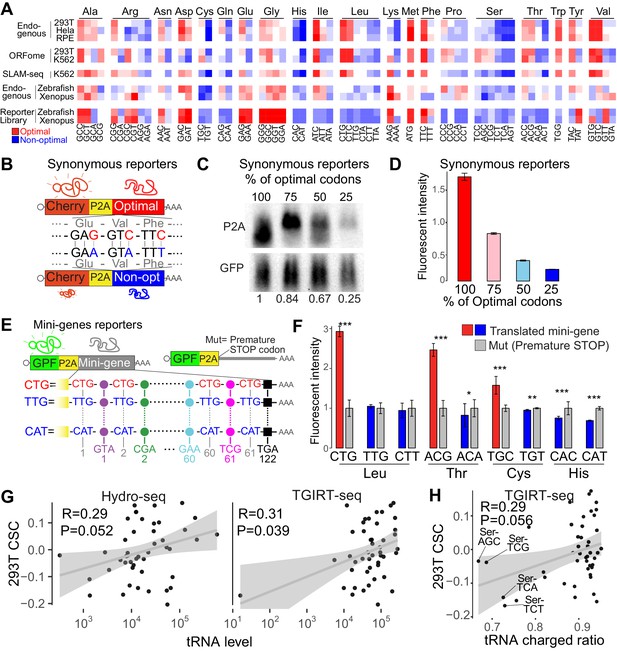
Synonymous codons affect mRNA stability in human lines.
(A) Heatmap showing the codon stability coefficient (CSC) sorted by the encoded amino-acid, for the indicated cell lines and approaches used to measure mRNA stability. The codon optimality scores calculated in zebrafish and Xenopus are also shown (Bazzini et al., 2016). (B) Diagram of the ‘Synonymous’ reporters. mRNAs differing only in synonymous mutations with different regulatory effects on mRNA stability. Each mRNA contains the coding region for mCherry fluorescent protein followed by a cis-acting hydrolase elements (P2A) and a coding region that different in the proportion of optimal and non-optimal codons but encoding for the same peptide (synonymous mutations). (C) Northern-blot showing that the synonymous reporter enriched in more optimal codons presented higher RNA level. GFP was used as internal control. Relative normalized quantifications are indicated. (D) The fluorescent intensity correlated with the proportion of optimal codon of each of the four reporter mRNAs differing only in synonymous codons. (E) Scheme of ‘Mini-gene’ reporters: all mini-genes mRNAs contain the coding region for Green fluorescent protein followed by a cis-acting hydrolase elements (P2A) and a coding region that contain the 61 different coding codons (colored circles) alternated with a single codon that give name to the mini-gene. The counterpart pair of each mini-gene (Mut) contains a single mutation in the P2A that prevent the coding region enriched in one particular codon to be translated. (F) Fluorescent GFP intensity of the indicated mini-genes normalized with internal control (mCherry). Unpaired t.test is used *p<0.05, **p<0.01, ***p<0.005. (G) Scatter plots showing the 293T CSC (endogenous mRNA) and tRNA levels quantified by Hydro-seq (Gogakos et al., 2017) and TGIRT-seq (Evans et al., 2017), Spearman rank correlations indicated. (H) Scatter plots showing the 293T CSC (endogenous mRNA) and tRNA charged ratios quantified by TGIRT-seq with periodate oxidation (Evans et al., 2017). The four nuclear tRNAs encoding for Serine are indicated. Spearman rank correlations indicated.
-
Figure 3—source data 1
Reporter sequences and oligos used for Figure 3.
- https://doi.org/10.7554/eLife.45396.012
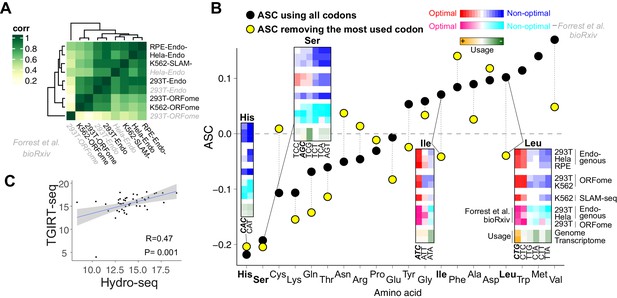
Few amino acids can potentially be related with mRNA stability in human lines.
(A) Heatmap showing the Spearman rank correlations between CSC calculated in different human cells lines and using different methods to calculate mRNA stability (Figure 1C) and scores from Forrest et al, preprint (bioRxiv_ (labeled in gray). For bioRxiv data (Forrest et al., 2018), the codon were ranked based on their optimality. (B) Scatter plot for ASC as the Person correlation coefficient between mRNA stability and amino acid occurrence (Bazzini et al., 2016) from 293T endogenous mRNA decay. The black dots indicate the ASC score considering all synonymous codons for a given amino acid, the yellow dots reflect the ASC score without taking in consideration the most abundant codon for each amino acid. Heatmap inside showing the CSC score from all our data and the CSC score rank from Forrest et al. (2018). The codon usages (genome and transcriptome) are also shown. (C) Scatter plot sowing the tRNA level measured with Hydro-seq and TGIRT-seq in 293T cells. The axis is log10 tRNA read counts. Spearman correlation was used, p value and R value indicated.
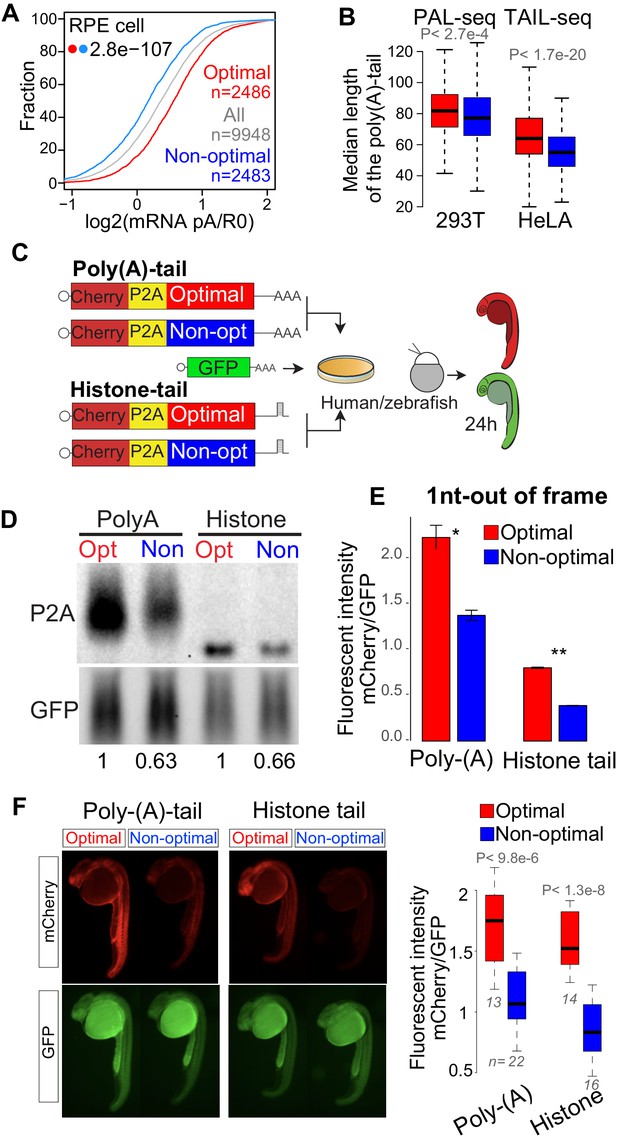
Poly(A)-tail is affected but not required for the codon optimality mechanism in human and zebrafish.
(A) Cumulative distribution of the poly(A) selected mRNA and total mRNA ratio in RPE cells showing that mRNA enriched in optimal codons present higher poly(A) ration than mRNA enriched in non-optimal codons, p-value indicated, Wilcoxon rank-sum test. (B) Box plots showing the length of the poly(A)-tails of mRNA enriched in optimal or non-optimal codons measured by PAL-seq (Subtelny et al., 2014) or TAIL-seq (Chang et al., 2014), the top 500 genes enriched in optimal or codons genes are used for each group, p-value indicated, Wilcoxon rank-sum test. (C) The 1nt-out of frame reporter containing either poly(A)- or histone-tail were transfected into human cells or co-injected into single-cell stage zebrafish embryos with a GFP construct as internal control. (D) Northern-blot showing that the 1nt-out of frame reporters enriched in optimal codons presents higher level of mRNA comparing to the counterpart enriched in non-optimal codon containing either a poly(A)- or histone- tail signal at 24 hr post-transfection. GFP was used as internal control. Relative normalized quantifications are indicated. (E) The 1nt-out of frame reporters enriched in optimal codons displayed higher fluorescent intensity than it counterparts enriched in non-optimal codons measure by cytometry, independent of the poly(A)- or histone-tail used. Unpaired t.test is used. *p<0.01, **p<0.005. (F) Fluorescence microscopy images of representative embryos at 24 hr post injection with the indicated 1nt-out of frame reporter and GFP. Box plot displays fluorescence quantification of 24 hr embryos injected with each reporter. mCherry fluorescence intensity was normalized to GFP intensity in each embryo, Poly-(A), n > 13 pairs, p = 9.3e-06 and Histone, n > 14 pairs, p = 1.3e-08, Wilcoxon rank-sum test.
-
Figure 4—source data 1
Reporter sequences and oligos used for Figure 4.
- https://doi.org/10.7554/eLife.45396.015
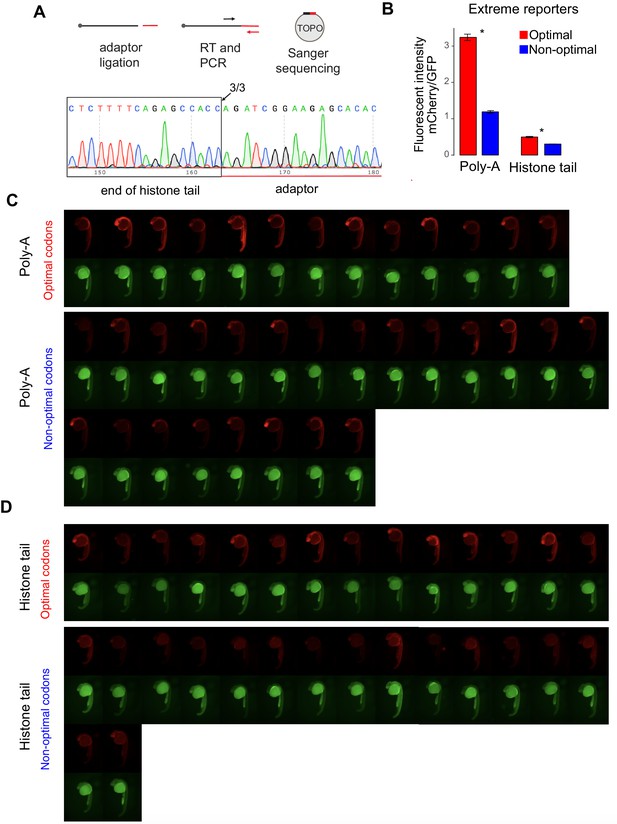
Poly(A)-tail is not required for the codon optimality mechanism in human and zebrafish.
(A) Diagram and sequencing result of the 3’RACE assay to detect end of histone tail mRNA. Total RNA was ligated with adaptor for qPCR at the 3’end. PCR product was inserted into TOPO vector for sanger sequencing. 3/3 colonies from the histone tail, showed sharp, precise end, indicating no poly(A)-tails. (B) The extreme reporter enriched in optimal codons displayed higher fluorescent intensity than the one enriched in non-optimal codons, independent of the poly(A)- or histone-tail used. unpaired t.test is used. *p<0.01 (C) Fluorescence microscopy images of embryos at 24 hr post injection with the indicated 1nt-out of frame reporter of poly(A)-tail and GFP. (D) Fluorescence microscopy images of embryos at 24 hr post injection with the indicated 1nt-out of frame reporter of histone-tail and GFP.
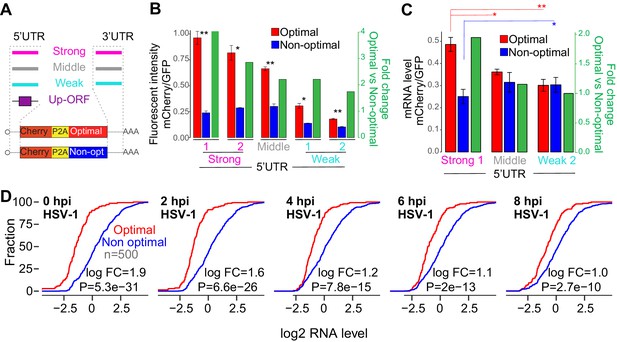
The number of ribosomes translating a mRNA modulates the codon-mediated effects on gene expression.
(A) Schematic representation of ‘1nt-out of frame’ reporters using different 5’UTR or 3’UTR or upstream ORFs (Up-ORF) driving different level of translation. (B) The fluorescent intensity correlated with the nature of the 5’UTR used as well as the fold change of fluorescent intensity between the reporter enriched in optimal and non-optimal codons. Unpaired t.test is used, *p < 1e-3, **p < 1e-4. (C) Reporters enriched in optimal codons displayed higher level of RNA when 5’UTR driving higher level of translation were used compared to constructs using 5’UTR driving lower level of translation. Contrary, the reporter enriched in non-optimal codons using the 5’UTR driving higher level of translation presented lower level of RNA than the one containing the weakest 5’UTR. Unpaired t.test is used, *p<0.05, **p<0.01. (D) Cumulative distribution of mRNA level from mRNAs enriched in either optimal or non-optimal codons during HSV-1 virus infection, p-value indicated, Wilcoxon rank-sum test.
-
Figure 5—source data 1
Reporter sequences and oligos used for Figure 5.
- https://doi.org/10.7554/eLife.45396.018
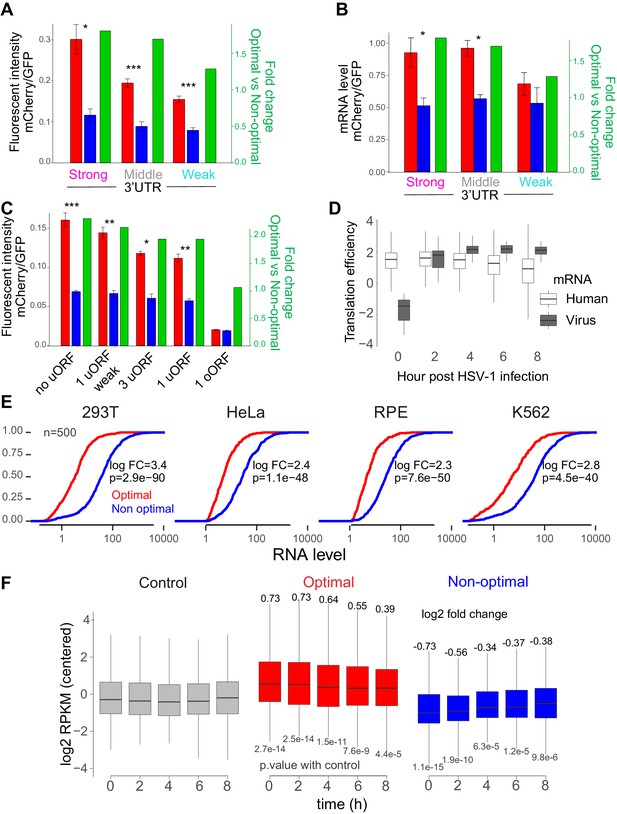
The number of ribosomes translating a mRNA modifies the codon-mediated effects on gene expression.
(A) The fluorescent intensity correlated with the nature of the 3’UTR used as well as the fold change between the reporter enriched in optimal and non-optimal codons. *p<0.05, **p<0.01, ***p<0.005, Unpaired t.test for all panels. (B) Reporters enriched in optimal codons displayed higher level of RNA when 3’UTR driving higher level of translation were used compared to constructs using the 3’UTR driving the lowest level of translation. The mRNA fold change between mRNA enriched in optimal and non-optimal was higher with 3’UTR driving higher level of translation than the one driving the lowest level. (C) The fluorescent intensity correlated with the nature of the upORFs used as well as the fold change between the reporter enriched in optimal and non-optimal codons. 5’UTR containing a weak up-stram-ORF (uORF), 1- and 3- canonical upORF were used. Overlapping ORF (oORF) with the mCherry was also used. (D) Boxplot showing the translation efficiency of both human host and virus mRNAs (Rutkowski et al., 2015). Human genes get decreased translation with time, virus genes get increased translation with time. (E) Cumulative distribution of mRNA level from top 500 mRNAs enriched in either optimal or non-optimal codons in 293T, HeLa, RPE, K562, P-values indicated, Wilcoxon rank-sum test. (F) Boxplot showing mRNA change after virus infection between top 500 mRNAs genes enriched in optimal or non-optimal codons. The data is centered (subtract the mean at each time point, so the mean expression is always at 0). As control group, a group of the same size of random genes not included in the category of optimal or not optimal was chosen. Log2 fold change and p.value compared to control for each time is indicated, calculated with a linear model.
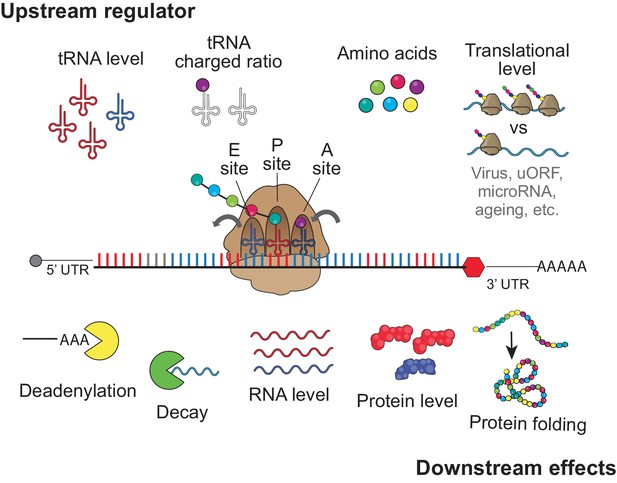
upstream regulator and downstream effect for codon optimality tRNA level, tRNA charged ratio, amino acid, and translational level might contribute to regulates the regulatory identity and/or strength of each codon to affect gene expression, by influencing the speed of translation elongation.
The downstream effects of the codon-meditated mechanism are RNA deadenylation, mRNA decay, mRNA level, protein level and protein folding.
Tables
| Reagent type | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line(Homo sapiens) | HEK293T | American Type Culture Collection | #CRL-11268 | |
| Cell line(H. sapiens) | Hela | American Type Culture Collection | #CCL2 | |
| Cell line(H. sapiens) | K562 | American Type Culture Collection | #CRL-4000 | |
| Cell line(H. sapiens) | RPE | American Type Culture Collection | #CCL-243, Lot #62995945 | |
| Recombinant DNA reagent | ORFome library | Sigma-Aldrich | TRC3 ORF Puromycin Arrayed Glycerol Library | |
| Commercial assay or kit | North2South Hybridization and Detection Kit | ThermoFisher | Cat#17097 | |
| Commercial assay or kit | SLAMseq Kinetics Kit | Lexogen | Cat#061.24 | |
| Software, algorithm | R for analysis and plot | version 3.5.1 |





